 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Bv1_005690_hemo.t1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Chenopodiaceae; Betoideae; Beta
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 846aa MW: 91613.4 Da PI: 6.3178 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 66.2 | 4.5e-21 | 147 | 202 | 1 | 56 |
TT--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 1 rrkRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
+++ +++t++q++eLe++F+++++p++++r eL+kkl L++rqVk+WFqNrR+++k
Bv1_005690_hemo.t1 147 KKRYHRHTPQQIQELESVFKECPHPDEKQRLELSKKLCLETRQVKFWFQNRRTQMK 202
688999***********************************************999 PP
| |||||||
| 2 | START | 191 | 5.7e-60 | 361 | 586 | 1 | 206 |
HHHHHHHHHHHHHHHHC-TT-EEEE.....EXCCTTEEEEEEESSS......SCEEEEEEEECCSCHHHHHHHHHCCCGGCT-TT-S.... CS
START 1 elaeeaaqelvkkalaeepgWvkss.....esengdevlqkfeeskv.....dsgealrasgvvdmvlallveellddkeqWdetla.... 77
ela+ a++elvk+a+ +ep+W + e++n +e+++++ +s + + ea+r++ +v+ ++ lve+l+d + +W e+++
Bv1_005690_hemo.t1 361 ELALGAMDELVKLAQTDEPLWLRNMdaggkEILNHEEYMRSINPSFGyksngFVPEASRETDMVIINSLALVETLMDAN-RWAEMFPgmia 450
57899*******************99****************9987799888999************************.*********** PP
EEEEEEEECTT......EEEEEEEEXXTTXX-SSX.EEEEEEEEEEE.TTS-EEEEEEEEE-TTS--.-TTSEE-EESSEEEEEEEECTCE CS
START 78 kaetlevissg......galqlmvaelqalsplvp.RdfvfvRyirqlgagdwvivdvSvdseqkppesssvvRaellpSgiliepksngh 161
+a+t++vissg g+lqlm aelq+lsplvp R + f+R+++q+ +g+w++vdvS+d ++ + s+ +v +++lpSg+++++++ng+
Bv1_005690_hemo.t1 451 RASTTDVISSGmggtrnGVLQLMHAELQVLSPLVPvRVVNFLRFCKQHAEGVWAVVDVSIDCIRESSGSPTYVNCRRLPSGCVVQDMPNGY 541
*****************************************************************999*********************** PP
EEEEEEE-EE--SSXXHHHHHHHHHHHHHHHHHHHHHHTXXXXXX CS
START 162 skvtwvehvdlkgrlphwllrslvksglaegaktwvatlqrqcek 206
skvtwveh++++++++h+l+r+l++ g+ +ga++w tlqrqc++
Bv1_005690_hemo.t1 542 SKVTWVEHAEYDESSIHQLYRPLISAGMGFGAQRWITTLQRQCQC 586
*******************************************85 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 1.45E-20 | 133 | 204 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.265 | 144 | 204 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 3.7E-18 | 145 | 208 | IPR001356 | Homeobox domain |
| Gene3D | G3DSA:1.10.10.60 | 7.1E-22 | 145 | 212 | IPR009057 | Homeodomain-like |
| CDD | cd00086 | 5.42E-19 | 146 | 204 | No hit | No description |
| Pfam | PF00046 | 1.3E-18 | 147 | 202 | IPR001356 | Homeobox domain |
| PROSITE pattern | PS00027 | 0 | 179 | 202 | IPR017970 | Homeobox, conserved site |
| PROSITE profile | PS50848 | 44.402 | 352 | 589 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 1.92E-31 | 354 | 586 | No hit | No description |
| CDD | cd08875 | 7.25E-117 | 356 | 585 | No hit | No description |
| SMART | SM00234 | 5.7E-44 | 361 | 586 | IPR002913 | START domain |
| Pfam | PF01852 | 4.5E-52 | 362 | 586 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 1.32E-24 | 614 | 839 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009827 | Biological Process | plant-type cell wall modification | ||||
| GO:0042335 | Biological Process | cuticle development | ||||
| GO:0043481 | Biological Process | anthocyanin accumulation in tissues in response to UV light | ||||
| GO:0048765 | Biological Process | root hair cell differentiation | ||||
| GO:0008289 | Molecular Function | lipid binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 846 aa Download sequence Send to blast |
MNFGGFLDSS GAGSDGGGSG GGASSARLVA DISYNHNHNN NNMSPAAISH SPHQHLQRLV 60 GPTSINKSFF NTPGLSLALQ TGMEGHGEVT KVTDQNYEGN NGNGGSGGSL GGVRRNREDE 120 HESRSGSDNL DGASGDDQDA ADLPPRKKRY HRHTPQQIQE LESVFKECPH PDEKQRLELS 180 KKLCLETRQV KFWFQNRRTQ MKTQLERHEN SILRQENDKL RAENMSIREA MRNPICTNCG 240 GQAMIGDISM EEQHLRIENA RLKDELDRVC TLASKFLGRP VSSLSTLTGS ISAPPMPSSS 300 LELAVGTNGF GPLTTVTTSS LPLAPDFGSG ISNALPVVSP TKSTANVSGI ERSFERSMYL 360 ELALGAMDEL VKLAQTDEPL WLRNMDAGGK EILNHEEYMR SINPSFGYKS NGFVPEASRE 420 TDMVIINSLA LVETLMDANR WAEMFPGMIA RASTTDVISS GMGGTRNGVL QLMHAELQVL 480 SPLVPVRVVN FLRFCKQHAE GVWAVVDVSI DCIRESSGSP TYVNCRRLPS GCVVQDMPNG 540 YSKVTWVEHA EYDESSIHQL YRPLISAGMG FGAQRWITTL QRQCQCLAIL MSSNIPSGDH 600 TAITASGRRS MLKLAQRMTN NFCAGVCAST VHKWNKLNVG NVDEDVQVMT RKSIDDPGEP 660 SGIVLSAATA VWLPVSPQRL FDFLRDERLR SEWDILSNGG PMQEMAHIAK GQDQGNCVSL 720 LRAGAMNPNQ SSMLILQETC TDASGSLVVY APVDIPAMHV VMNGGDSAYV ALLPSGFAIT 780 PDGPSTRGPN GNAGPQRVGG SLLSVAFQIL VNSLPTAKLT VESVETVNNL ISCTVQKIKA 840 ALNCES |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor involved in the regulation of the tissue-specific accumulation of anthocyanins and in cellular organization of the primary root. {ECO:0000269|PubMed:10402424}. | |||||
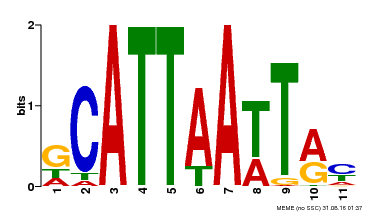
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00421 | DAP | Transfer from AT4G00730 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010671720.1 | 0.0 | PREDICTED: homeobox-leucine zipper protein ANTHOCYANINLESS 2 | ||||
| Swissprot | Q0WV12 | 0.0 | ANL2_ARATH; Homeobox-leucine zipper protein ANTHOCYANINLESS 2 | ||||
| TrEMBL | A0A0K9QLI3 | 0.0 | A0A0K9QLI3_SPIOL; Uncharacterized protein | ||||
| STRING | XP_010671720.1 | 0.0 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G00730.1 | 0.0 | HD-ZIP family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



