 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Bv1_009470_kyxu.t1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Chenopodiaceae; Betoideae; Beta
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 383aa MW: 41581 Da PI: 5.8579 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 65.2 | 1.3e-20 | 175 | 224 | 2 | 55 |
AP2 2 gykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
y+GVr+++ +g+WvAeIr p++ r+r +lg+f+taeeAa a+++a+ kl+g
Bv1_009470_kyxu.t1 175 LYRGVRQRH-WGKWVAEIRLPKN---RTRLWLGTFDTAEEAALAYDKAAYKLRG 224
59******9.**********965...5*************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00018 | 1.08E-33 | 174 | 234 | No hit | No description |
| PROSITE profile | PS51032 | 23.143 | 175 | 232 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 5.4E-38 | 175 | 238 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 3.8E-32 | 175 | 233 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 4.05E-23 | 175 | 233 | IPR016177 | DNA-binding domain |
| Pfam | PF00847 | 4.0E-14 | 176 | 224 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 3.9E-11 | 176 | 187 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 3.9E-11 | 198 | 214 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0009611 | Biological Process | response to wounding | ||||
| GO:0009736 | Biological Process | cytokinin-activated signaling pathway | ||||
| GO:0009873 | Biological Process | ethylene-activated signaling pathway | ||||
| GO:0010017 | Biological Process | red or far-red light signaling pathway | ||||
| GO:0045595 | Biological Process | regulation of cell differentiation | ||||
| GO:0071472 | Biological Process | cellular response to salt stress | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 383 aa Download sequence Send to blast |
MATTMDMIYS SSSDCAGCLD PLGNELMEAL SPFMKTSPSS SPPPSSSLST LPEHSTISST 60 STSTTQSYSS TSFSLSSFPQ DPTSFFQDGY CYSQQAPFFG LNHPDPSTSL GLNNLPQAQF 120 NYPIAQFNYP QAQQQVTYSY PPPLPSYQST NMNSFLSPKP IPMKQSGTPP KPTKLYRGVR 180 QRHWGKWVAE IRLPKNRTRL WLGTFDTAEE AALAYDKAAY KLRGDFARLN FPNLRHEGAH 240 IGGEFGEYKP LHSSVNAKLE AICESLAKQG NEKHQGKSGK SKKTKGVVTV TSSSSSSSCS 300 SSLVAAASSS NDDQPQQQQQ QQQQMPDVVK AECASDSEVG SGGSSPLSDL TFGDADDLGV 360 ENLNYLLEKC PSHEIDWDAL LSS |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1gcc_A | 2e-22 | 174 | 232 | 1 | 60 | ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00026 | PBM | Transfer from AT1G78080 | Download |
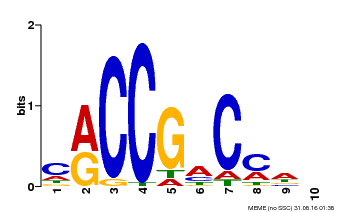
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010676915.1 | 0.0 | PREDICTED: ethylene-responsive transcription factor RAP2-4 | ||||
| TrEMBL | A0A088FRQ7 | 1e-151 | A0A088FRQ7_SUASA; DREBb | ||||
| STRING | XP_010676915.1 | 0.0 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G78080.1 | 3e-63 | related to AP2 4 | ||||



