 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Bv2_045740_qaam.t1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Chenopodiaceae; Betoideae; Beta
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 399aa MW: 44695.6 Da PI: 9.0119 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 101.6 | 5.1e-32 | 85 | 138 | 2 | 55 |
G2-like 2 prlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
prlrWtp+LH +Fv+ave+LGG+e+AtPk +l+lm++kgL+++hvkSHLQ+YR+
Bv2_045740_qaam.t1 85 PRLRWTPDLHLCFVHAVERLGGQERATPKLVLQLMNIKGLSIAHVKSHLQMYRS 138
8****************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 12.176 | 81 | 141 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 3.6E-30 | 81 | 139 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 2.33E-14 | 83 | 139 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 1.1E-22 | 85 | 140 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 4.4E-9 | 86 | 137 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 399 aa Download sequence Send to blast |
MIRAGSGSTE GSKTSLSNDI IDDEFDEDDN KVEEEEVDDN SNNTNSKVVI KINGSSSSSS 60 TVEEDEKKRT TSSGSVRPYN RSKMPRLRWT PDLHLCFVHA VERLGGQERA TPKLVLQLMN 120 IKGLSIAHVK SHLQMYRSKK IDESNQEAMS NNQGIFSDGR DNIYNLSQLP MLQGYNHQRS 180 PFNLRYGDPS WRGHSNQIYG PYNVGTSSDR PRNGSNFNSL SERMIFSSNS INSQVTNVNF 240 LQGQLSNWRT QQSLKNTPIF QPHGSSSSQS ISRPSINLAI SPTTEVAPEK REKSNSFNFL 300 KPSNNNNIEI KQRTIVNHLK RKSLDLDLSL ALAPKNEVDH SLTKANSKDK EKDCELLSLS 360 LLHTPPFSSC SSDKFINKLK EEDASSKNKQ AKTSLDLTL |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4k_A | 2e-18 | 86 | 140 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4k_B | 2e-18 | 86 | 140 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_A | 2e-18 | 86 | 140 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_B | 2e-18 | 86 | 140 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_C | 2e-18 | 86 | 140 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_D | 2e-18 | 86 | 140 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_A | 2e-18 | 86 | 140 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_C | 2e-18 | 86 | 140 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_D | 2e-18 | 86 | 140 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_F | 2e-18 | 86 | 140 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_H | 2e-18 | 86 | 140 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_J | 2e-18 | 86 | 140 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00297 | DAP | Transfer from AT2G38300 | Download |
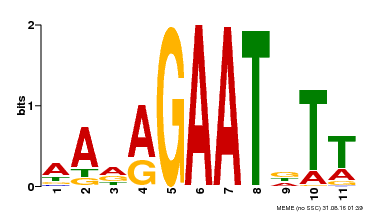
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010693076.1 | 0.0 | PREDICTED: two-component response regulator ARR10 | ||||
| TrEMBL | A0A0J8BEJ5 | 0.0 | A0A0J8BEJ5_BETVU; Uncharacterized protein | ||||
| STRING | XP_010693076.1 | 0.0 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G38300.1 | 7e-51 | G2-like family protein | ||||



