 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Bv2_046230_sgnn.t1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Chenopodiaceae; Betoideae; Beta
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 336aa MW: 38854.9 Da PI: 7.8674 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 160.1 | 8.5e-50 | 37 | 166 | 3 | 128 |
NAM 3 pGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLp.kkvkaeekewyfFskrdkkyatgkrknratksgyWkatgkdkev 92
pGfrFhPtdeelv++yLk+kve+++ e+ik++diyk++PwdL ++ + ++kewyf++kr +ky+++ r+nr+t sg+Wkatg d+++
Bv2_046230_sgnn.t1 37 PGFRFHPTDEELVSFYLKRKVEKRRPLSIEIIKQIDIYKYDPWDLAkASSSIGDKEWYFYCKRGRKYRNSIRPNRVTGSGFWKATGIDRPI 127
9**********************99555599**************844566699************************************* PP
NAM 93 lsk...kgelvglkktLvfykgrapkgektdWvmheyrl 128
+++ +++ +glkk+Lv+y+g+a kg+ktdW+mhe+rl
Bv2_046230_sgnn.t1 128 YCNegeGNQCIGLKKSLVYYRGSAGKGTKTDWMMHEFRL 166
**97766677***************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101941 | 4.32E-53 | 34 | 200 | IPR003441 | NAC domain |
| PROSITE profile | PS51005 | 53.733 | 35 | 200 | IPR003441 | NAC domain |
| Pfam | PF02365 | 7.4E-25 | 37 | 166 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005992 | Biological Process | trehalose biosynthetic process | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0006561 | Biological Process | proline biosynthetic process | ||||
| GO:0009718 | Biological Process | anthocyanin-containing compound biosynthetic process | ||||
| GO:0010120 | Biological Process | camalexin biosynthetic process | ||||
| GO:0042538 | Biological Process | hyperosmotic salinity response | ||||
| GO:1900056 | Biological Process | negative regulation of leaf senescence | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 336 aa Download sequence Send to blast |
MEVKRCNSQD DQQERRDSKI SGGGLVDGVD DDEGPLPGFR FHPTDEELVS FYLKRKVEKR 60 RPLSIEIIKQ IDIYKYDPWD LAKASSSIGD KEWYFYCKRG RKYRNSIRPN RVTGSGFWKA 120 TGIDRPIYCN EGEGNQCIGL KKSLVYYRGS AGKGTKTDWM MHEFRLPTDQ HQHDQAANFS 180 KPKDTAPEAE IWTLCRIFKR NKRHTPDWRE LSSRKNAVNS SSKTSSIDSN TNQAYYISFG 240 THEVQPNSSY SHLHVVDDEN NNIYLGNNLN ITTCDQKNVN FQPEYQQQWN SLAHTPSSTT 300 RSSMNNENDL QSFAYYGHNW DDLNSVVEYA VNKPFL |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 3ulx_A | 4e-44 | 37 | 198 | 17 | 166 | Stress-induced transcription factor NAC1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
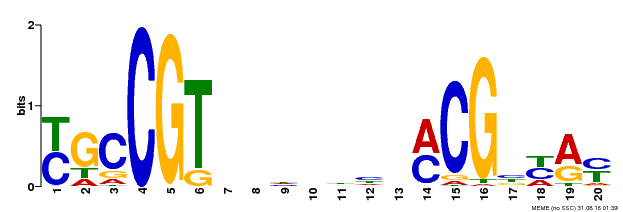
| UniProt | Transcription factor that binds to the 5'- RRYGCCGT-3' consensus core sequence. Central longevity regulator. Negative regulator of leaf senescence. Modulates cellular H(2)O(2) levels and enhances tolerance to various abiotic stresses through the regulation of DREB2A. {ECO:0000269|PubMed:22345491}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00310 | DAP | Transfer from AT2G43000 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Up-regulated by H(2)O(2), paraquat, ozone, 3-aminotriazole and salt stress. {ECO:0000269|PubMed:22345491}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010693169.1 | 0.0 | PREDICTED: transcription factor JUNGBRUNNEN 1-like | ||||
| Swissprot | Q9SK55 | 7e-84 | NAC42_ARATH; Transcription factor JUNGBRUNNEN 1 | ||||
| TrEMBL | A0A0J8BD73 | 0.0 | A0A0J8BD73_BETVU; Uncharacterized protein | ||||
| STRING | XP_010693169.1 | 0.0 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G12910.1 | 2e-76 | NAC family protein | ||||



