 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Bv3_060800_jzfr.t1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Chenopodiaceae; Betoideae; Beta
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 412aa MW: 46395.4 Da PI: 7.2041 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 108.3 | 4.2e-34 | 35 | 89 | 1 | 55 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
kprl+Wtp+LHerFveav+qLGG++kAtPkt+++lm+++gLtl+h+kSHLQkYRl
Bv3_060800_jzfr.t1 35 KPRLKWTPDLHERFVEAVNQLGGADKATPKTVMKLMGIPGLTLYHLKSHLQKYRL 89
79****************************************************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 10.849 | 32 | 92 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 4.3E-32 | 33 | 90 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 4.84E-17 | 34 | 90 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 3.5E-23 | 35 | 90 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 6.4E-10 | 37 | 88 | IPR001005 | SANT/Myb domain |
| Pfam | PF14379 | 1.9E-9 | 134 | 155 | IPR025756 | MYB-CC type transcription factor, LHEQLE-containing domain |
| Pfam | PF14379 | 5.9E-10 | 194 | 219 | IPR025756 | MYB-CC type transcription factor, LHEQLE-containing domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 412 aa Download sequence Send to blast |
MYPSPRISAP QERQLFLQGG NGSRDSNLVL STDAKPRLKW TPDLHERFVE AVNQLGGADK 60 ATPKTVMKLM GIPGLTLYHL KSHLQKYRLS KNIHGQTSSG ANKTVVMSSD KMQETSPSHT 120 SSINIAHQPN KNLHINEALQ MQIEVQRRLH EQLEVLDPVY FQFKQNGLSP KAETTTAAQN 180 LQRHHPKNSR DKDVQRRLQL RIEAQGKYLQ AVLEKAQETL GKQNIGVVGL EAAKLQLSEL 240 VSKVSNQCLN LAFAEMAEEQ QGLCPKQQIQ PHKPMGDCSL DSCLTSCDGP QLREQDMHNS 300 ELGLRPYNDT RFLEGNEFNE NLKEHATILF PTTTQQDTAK MMFRMEGACS DLSMSVGIRR 360 DTRDELEQST FQLDNKNYKL GSFSGELDLN ARDEPDVPST IRHLDLNGLS WT |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4k_A | 2e-21 | 34 | 91 | 1 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4k_B | 2e-21 | 34 | 91 | 1 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_A | 2e-21 | 35 | 91 | 1 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_B | 2e-21 | 35 | 91 | 1 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_C | 2e-21 | 35 | 91 | 1 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_D | 2e-21 | 35 | 91 | 1 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_A | 2e-21 | 34 | 91 | 1 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_C | 2e-21 | 34 | 91 | 1 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_D | 2e-21 | 34 | 91 | 1 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_F | 2e-21 | 34 | 91 | 1 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_H | 2e-21 | 34 | 91 | 1 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_J | 2e-21 | 34 | 91 | 1 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00331 | DAP | Transfer from AT3G04030 | Download |
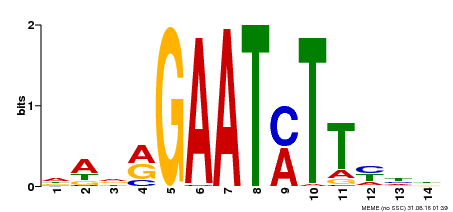
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010672721.1 | 0.0 | PREDICTED: myb-related protein 2-like isoform X2 | ||||
| TrEMBL | A0A0K9RSG6 | 0.0 | A0A0K9RSG6_SPIOL; Uncharacterized protein | ||||
| STRING | XP_010672720.1 | 0.0 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G04030.3 | 1e-109 | G2-like family protein | ||||



