 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Bv3_064050_dwac.t1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Chenopodiaceae; Betoideae; Beta
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 373aa MW: 43573.4 Da PI: 8.1856 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 12.9 | 0.00034 | 21 | 43 | 1 | 23 |
EEETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCpdCgksFsrksnLkrHirtH 23
y C+ Cg + s ks+L++Hi +H
Bv3_064050_dwac.t1 21 YYCEFCGICRSKKSLLTSHILSH 43
89*******************96 PP
| |||||||
| 2 | zf-C2H2 | 21.5 | 6.1e-07 | 69 | 90 | 2 | 23 |
EETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 2 kCpdCgksFsrksnLkrHirtH 23
+C+ Cg +F+ + +Lk+H+ +H
Bv3_064050_dwac.t1 69 TCEECGATFKKPAHLKQHMLSH 90
6*******************99 PP
| |||||||
| 3 | zf-C2H2 | 21.6 | 5.7e-07 | 96 | 120 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirtH 23
++Cp dC++s++rk++L+rH+ H
Bv3_064050_dwac.t1 96 FTCPvdDCNTSYRRKDHLNRHLLQH 120
89*******************9887 PP
| |||||||
| 4 | zf-C2H2 | 15.1 | 6.5e-05 | 125 | 150 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt.H 23
+kCp +C+k F+ + n rH++ H
Bv3_064050_dwac.t1 125 FKCPveNCTKEFTVHGNISRHMKEfH 150
89*********************988 PP
| |||||||
| 5 | zf-C2H2 | 18.4 | 6e-06 | 166 | 190 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirtH 23
++C+ Cgk F+ s+L++H +H
Bv3_064050_dwac.t1 166 HVCQeeGCGKEFKYASQLRKHAESH 190
789999***************9988 PP
| |||||||
| 6 | zf-C2H2 | 15.4 | 5.3e-05 | 255 | 279 | 2 | 23 |
EET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 2 kCp..dCgksFsrksnLkrHirt.H 23
kC+ C+ sF++ksn+++H + H
Bv3_064050_dwac.t1 255 KCSfeGCNHSFTTKSNMQQHVKAvH 279
79999***************99877 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50157 | 8.871 | 21 | 49 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 13 | 21 | 43 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 11.801 | 68 | 95 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.0041 | 68 | 90 | IPR015880 | Zinc finger, C2H2-like |
| Gene3D | G3DSA:3.30.160.60 | 6.6E-5 | 69 | 86 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE pattern | PS00028 | 0 | 70 | 90 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 1.89E-10 | 76 | 128 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 2.8E-12 | 87 | 122 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 9.91 | 96 | 125 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 6.1E-4 | 96 | 120 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 98 | 120 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 9.56E-6 | 123 | 152 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 2.1E-5 | 123 | 147 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 10.325 | 125 | 155 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.019 | 125 | 150 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 127 | 150 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 6.9E-7 | 161 | 190 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 10.658 | 166 | 195 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.004 | 166 | 190 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 168 | 190 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 2.6 | 197 | 222 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 199 | 222 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 27 | 225 | 246 | IPR015880 | Zinc finger, C2H2-like |
| SuperFamily | SSF57667 | 7.01E-8 | 237 | 292 | No hit | No description |
| PROSITE profile | PS50157 | 11.697 | 254 | 284 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 2.0E-4 | 254 | 279 | IPR015880 | Zinc finger, C2H2-like |
| Gene3D | G3DSA:3.30.160.60 | 1.0E-7 | 255 | 279 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE pattern | PS00028 | 0 | 256 | 279 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 1.1E-4 | 280 | 308 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 5.2 | 285 | 311 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 287 | 311 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005730 | Cellular Component | nucleolus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008097 | Molecular Function | 5S rRNA binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| GO:0080084 | Molecular Function | 5S rDNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 373 aa Download sequence Send to blast |
MGEEHEESSK IGPIFRDIRR YYCEFCGICR SKKSLLTSHI LSHHQNEMEK RREIEEEEEE 60 GEEKKCNNTC EECGATFKKP AHLKQHMLSH SVERLFTCPV DDCNTSYRRK DHLNRHLLQH 120 QGKLFKCPVE NCTKEFTVHG NISRHMKEFH EHKPAVPDNG KEQSKHVCQE EGCGKEFKYA 180 SQLRKHAESH VLQSSETFCG DPGCMKPFAS VECLRAHMRS CHQYVNCEVC GSKHLRKNYK 240 RHLQTHDEHL PTTLKCSFEG CNHSFTTKSN MQQHVKAVHL KLKPFICSIP GCGMRFAFKH 300 VRDNHEKTSR HVYVHGDLEE FDEQFRSRPR GGQKRKLPDI EMLMRKRISA PNHSDSAQNR 360 SSEFLMWLLS DDT |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1tf6_A | 9e-15 | 72 | 217 | 19 | 155 | PROTEIN (TRANSCRIPTION FACTOR IIIA) |
| 1tf6_D | 9e-15 | 72 | 217 | 19 | 155 | PROTEIN (TRANSCRIPTION FACTOR IIIA) |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Essential protein (PubMed:22353599). Isoform 1 is a transcription activator the binds both 5S rDNA and 5S rRNA and stimulates the transcription of 5S rRNA gene (PubMed:12711688, PubMed:22353599). Isoform 1 regulates 5S rRNA levels during development (PubMed:22353599). {ECO:0000269|PubMed:12711688, ECO:0000269|PubMed:22353599}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00229 | DAP | Transfer from AT1G72050 | Download |
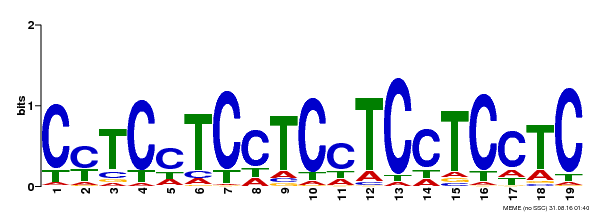
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010673198.1 | 0.0 | PREDICTED: transcription factor IIIA | ||||
| Swissprot | Q84MZ4 | 1e-138 | TF3A_ARATH; Transcription factor IIIA | ||||
| TrEMBL | A0A0K9R4E8 | 0.0 | A0A0K9R4E8_SPIOL; Uncharacterized protein | ||||
| STRING | XP_010673198.1 | 0.0 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G72050.2 | 1e-129 | transcription factor IIIA | ||||



