 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Bv3_064050_dwac.t3 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Chenopodiaceae; Betoideae; Beta
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 228aa MW: 26434 Da PI: 8.6856 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 19.3 | 3.1e-06 | 21 | 45 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirtH 23
++C+ Cgk F+ s+L++H +H
Bv3_064050_dwac.t3 21 HVCQeeGCGKEFKYASQLRKHAESH 45
789999***************9988 PP
| |||||||
| 2 | zf-C2H2 | 16.3 | 2.8e-05 | 110 | 134 | 2 | 23 |
EET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 2 kCp..dCgksFsrksnLkrHirt.H 23
kC+ C+ sF++ksn+++H + H
Bv3_064050_dwac.t3 110 KCSfeGCNHSFTTKSNMQQHVKAvH 134
79999***************99877 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:3.30.160.60 | 3.5E-7 | 16 | 45 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 0.004 | 21 | 45 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 10.658 | 21 | 50 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 23 | 45 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 2.6 | 52 | 77 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 54 | 77 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 27 | 80 | 101 | IPR015880 | Zinc finger, C2H2-like |
| SuperFamily | SSF57667 | 4.59E-8 | 92 | 147 | No hit | No description |
| SMART | SM00355 | 2.0E-4 | 109 | 134 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 11.697 | 109 | 139 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 5.2E-8 | 110 | 134 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE pattern | PS00028 | 0 | 111 | 134 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 7.3E-5 | 135 | 163 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 5.2 | 140 | 166 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 142 | 166 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005730 | Cellular Component | nucleolus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008097 | Molecular Function | 5S rRNA binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| GO:0080084 | Molecular Function | 5S rDNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 228 aa Download sequence Send to blast |
MKEFHEHKPA VPDNGKEQSK HVCQEEGCGK EFKYASQLRK HAESHVLQSS ETFCGDPGCM 60 KPFASVECLR AHMRSCHQYV NCEVCGSKHL RKNYKRHLQT HDEHLPTTLK CSFEGCNHSF 120 TTKSNMQQHV KAVHLKLKPF ICSIPGCGMR FAFKHVRDNH EKTSRHVYVH GDLEEFDEQF 180 RSRPRGGQKR KLPDIEMLMR KRISAPNHSD SAQNRSSEFL MWLLSDDT |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2gli_A | 4e-11 | 23 | 171 | 4 | 155 | PROTEIN (FIVE-FINGER GLI) |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Essential protein (PubMed:22353599). Isoform 1 is a transcription activator the binds both 5S rDNA and 5S rRNA and stimulates the transcription of 5S rRNA gene (PubMed:12711688, PubMed:22353599). Isoform 1 regulates 5S rRNA levels during development (PubMed:22353599). {ECO:0000269|PubMed:12711688, ECO:0000269|PubMed:22353599}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00229 | DAP | Transfer from AT1G72050 | Download |
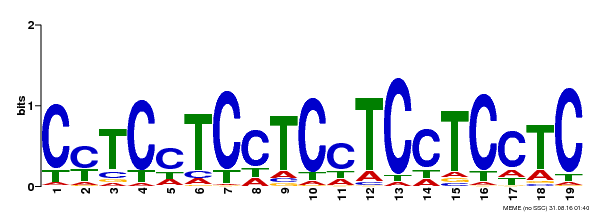
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010673198.1 | 1e-169 | PREDICTED: transcription factor IIIA | ||||
| Swissprot | Q84MZ4 | 3e-79 | TF3A_ARATH; Transcription factor IIIA | ||||
| TrEMBL | A0A0K9R4E8 | 1e-134 | A0A0K9R4E8_SPIOL; Uncharacterized protein | ||||
| STRING | XP_010673198.1 | 1e-169 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G72050.1 | 3e-82 | transcription factor IIIA | ||||



