 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Bv3_069990_sghj.t1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Chenopodiaceae; Betoideae; Beta
|
||||||||
| Family | CPP | ||||||||
| Protein Properties | Length: 761aa MW: 83178 Da PI: 7.0937 | ||||||||
| Description | CPP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | TCR | 51.1 | 2.7e-16 | 466 | 504 | 2 | 40 |
TCR 2 ekkgCnCkkskClkkYCeCfaagkkCseeCkCedCkNke 40
++k+CnCkkskClk+YCeCfaag++C e C+C+dC+Nk
Bv3_069990_sghj.t1 466 SCKRCNCKKSKCLKLYCECFAAGVYCVEPCSCQDCFNKP 504
689**********************************96 PP
| |||||||
| 2 | TCR | 50.9 | 3.1e-16 | 551 | 589 | 1 | 39 |
TCR 1 kekkgCnCkkskClkkYCeCfaagkkCseeCkCedCkNk 39
++k+gCnCkks+ClkkYCeC++ g+ Cs +C+Ce+CkN
Bv3_069990_sghj.t1 551 RHKRGCNCKKSNCLKKYCECYQGGVGCSINCRCEGCKNA 589
589***********************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM01114 | 1.0E-17 | 465 | 506 | IPR033467 | Tesmin/TSO1-like CXC domain |
| PROSITE profile | PS51634 | 37.785 | 466 | 591 | IPR005172 | CRC domain |
| Pfam | PF03638 | 8.7E-12 | 468 | 503 | IPR005172 | CRC domain |
| SMART | SM01114 | 3.7E-17 | 551 | 592 | IPR033467 | Tesmin/TSO1-like CXC domain |
| Pfam | PF03638 | 1.2E-11 | 553 | 589 | IPR005172 | CRC domain |
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 761 aa Download sequence Send to blast |
MDTPERNQIG TPISKMEESP FSSFINNLSP INPTKATHVS QTFSSLSFTS PPSVFTSPHV 60 SLQKDYKFLR RHQVSDLAKS PFALEAGNKD SVAKEVVSSG HLSDSLEIVQ VRGETASGSE 120 DSAKPLEENT DCAKELPRVL KYDIGSPRCK TAVTSSGNET NCEPDASAAL VQYVQEPSVR 180 GSFGTELNSQ VMNHDNEETC DWQSLISGSS DLFIFDSPNI ATSKESVQKS EEKETNVFTS 240 LLSRFPHDEA FSMQTTLHDG SSNCNEAYES IENSTQPTEA PELKQMEQPP ETLEISSVNE 300 LLCGDSVGKK GKQPFSNLHR GLRRRCLTFE IPRTKKSIDS ALFSSSLSSL QSEEKPLSCI 360 RQPITPGNDS SRRILPGIGL HLNALAAASK DCSNPNQEII SPTAQDPFMK SLDSSSLRKD 420 EGPAENVAEI AENSSQASVY AAIEELNPNS PKKKRRKFET IGEGESCKRC NCKKSKCLKL 480 YCECFAAGVY CVEPCSCQDC FNKPIHEETV LATRKQIESR NPLAFAPKVI RTTDSVQELG 540 EDTTRTPASA RHKRGCNCKK SNCLKKYCEC YQGGVGCSIN CRCEGCKNAF GRKDGSTMIA 600 TEAEIEKLQA SEKSIMDKSS DIHRSHDEDD QHSEFSIPQT PSRLYRPLVP LATSSKGKPP 660 RSSLAMSGLS SSLHPVDGPG RSTFLRSQSC AEKHHHSVSV DEIPAVLQNT SPPSSGLKTL 720 SPNSKRVLSP HNANGQSHGL RSSRKLILQS IPAFPSLSPR R |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5fd3_A | 1e-16 | 470 | 588 | 14 | 120 | Protein lin-54 homolog |
| 5fd3_B | 1e-16 | 470 | 588 | 14 | 120 | Protein lin-54 homolog |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 451 | 455 | KKKRR |
| 2 | 451 | 456 | KKKRRK |
| 3 | 452 | 456 | KKRRK |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00624 | PBM | Transfer from PK22848.1 | Download |
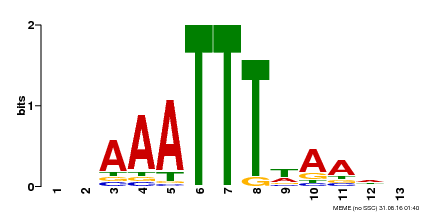
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010694035.1 | 0.0 | PREDICTED: protein tesmin/TSO1-like CXC 2 | ||||
| TrEMBL | A0A0K9R5V1 | 0.0 | A0A0K9R5V1_SPIOL; Uncharacterized protein | ||||
| STRING | XP_010694035.1 | 0.0 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G14770.1 | 1e-117 | TESMIN/TSO1-like CXC 2 | ||||



