 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Bv4_079320_gsmr.t1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Chenopodiaceae; Betoideae; Beta
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 683aa MW: 73844.5 Da PI: 6.9389 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 50 | 5.1e-16 | 443 | 489 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
hn ErrRRdriN+++ L+el+P++ K +Ka++L +A+eY+k Lq
Bv4_079320_gsmr.t1 443 VHNLSERRRRDRINEKMRALQELIPNC-----NKADKASMLDEAIEYLKTLQ 489
6*************************8.....6******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00083 | 2.77E-17 | 435 | 493 | No hit | No description |
| Gene3D | G3DSA:4.10.280.10 | 4.8E-20 | 436 | 497 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 1.57E-20 | 436 | 502 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 18.228 | 439 | 488 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 2.3E-13 | 443 | 489 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 2.7E-17 | 445 | 494 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009704 | Biological Process | de-etiolation | ||||
| GO:0009740 | Biological Process | gibberellic acid mediated signaling pathway | ||||
| GO:0010017 | Biological Process | red or far-red light signaling pathway | ||||
| GO:0031539 | Biological Process | positive regulation of anthocyanin metabolic process | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 683 aa Download sequence Send to blast |
MPLTELVRMF KGKVEVNQDG VSNSIDPSSR PNTDCVELVW ENGQLMMQGK TRKSSNSNNF 60 QTVGPKLQDG RNVGNFSRMT KFGTMDSLMA DYPLSVPSCE MGLDQDVDLV PWLNYPMEEH 120 LHNDYTSDFL HELSGVTIND LSTENHLPYR ERKSTFHGDG HDTSVSLKHG HTSKASSSFE 180 KDNRGGSSEV PQFMFPFQQS IPRSGISDII GNSAGNAHHV ASRDSANNPS SLSAFTSLRL 240 QKLDAGQPST SSGFTNFPYF SRPTGLSRAN QVTVNPSKRL ENIDKRCTLN SSNPANSTLV 300 NMDSGYSQKD IVSHNQSIMT PANRDSEALP AKPTQEPRSA DQPMDREDIA KNHVSKNQAC 360 DAPMSKVGTS GDRNMEPVVA TSSVCSANSA ERTSDEPARN LKRKSHDTTE SEGPSEEAEE 420 ESVGARKAGH GRNGSKRSRA AEVHNLSERR RRDRINEKMR ALQELIPNCN KADKASMLDE 480 AIEYLKTLQI QVQILSMGAG LYMQPMMLPP GMQPIHGAHM PHFSPMGLGM GMGMGFGMNM 540 LNMNNGPKML PFQGPHYPVP GTGFQGMPGS NLPAFPHPGQ VLPMSMQQAP IVPPPISGSF 600 LNMPIGLPAS GVAGPSSAPQ LAPPGSKKDA NPHGLSSMAS NNVKSSLNPA TIQGSNEGNQ 660 VCHDQTQAVN AVETASGKDA MHS |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 401 | 405 | KRKSH |
| 2 | 447 | 452 | ERRRRD |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00081 | ChIP-seq | Transfer from AT1G09530 | Download |
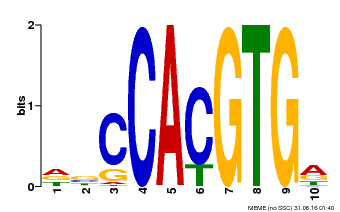
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_019104506.1 | 0.0 | PREDICTED: transcription factor PIF3 isoform X1 | ||||
| Refseq | XP_019104507.1 | 0.0 | PREDICTED: transcription factor PIF3 isoform X1 | ||||
| Refseq | XP_019104508.1 | 0.0 | PREDICTED: transcription factor PIF3 isoform X1 | ||||
| TrEMBL | A0A0K9RD81 | 0.0 | A0A0K9RD81_SPIOL; Uncharacterized protein | ||||
| STRING | XP_010674470.1 | 0.0 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G09530.2 | 7e-37 | phytochrome interacting factor 3 | ||||



