 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Bv4_079320_gsmr.t2 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Chenopodiaceae; Betoideae; Beta
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 675aa MW: 72904.4 Da PI: 6.9387 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 50.1 | 5.1e-16 | 435 | 481 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
hn ErrRRdriN+++ L+el+P++ K +Ka++L +A+eY+k Lq
Bv4_079320_gsmr.t2 435 VHNLSERRRRDRINEKMRALQELIPNC-----NKADKASMLDEAIEYLKTLQ 481
6*************************8.....6******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00083 | 2.70E-17 | 427 | 485 | No hit | No description |
| Gene3D | G3DSA:4.10.280.10 | 4.8E-20 | 428 | 489 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 1.57E-20 | 428 | 494 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 18.228 | 431 | 480 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 2.2E-13 | 435 | 481 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 2.7E-17 | 437 | 486 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009704 | Biological Process | de-etiolation | ||||
| GO:0009740 | Biological Process | gibberellic acid mediated signaling pathway | ||||
| GO:0010017 | Biological Process | red or far-red light signaling pathway | ||||
| GO:0031539 | Biological Process | positive regulation of anthocyanin metabolic process | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 675 aa Download sequence Send to blast |
MFKGKVEVNQ DGVSNSIDPS SRPNTDCVEL VWENGQLMMQ GKTRKSSNSN NFQTVGPKLQ 60 DGRNVGNFSR MTKFGTMDSL MADYPLSVPS CEMGLDQDVD LVPWLNYPME EHLHNDYTSD 120 FLHELSGVTI NDLSTENHLP YRERKSTFHG DGHDTSVSLK HGHTSKASSS FEKDNRGGSS 180 EVPQFMFPFQ QSIPRSGISD IIGNSAGNAH HVASRDSANN PSSLSAFTSL RLQKLDAGQP 240 STSSGFTNFP YFSRPTGLSR ANQVTVNPSK RLENIDKRCT LNSSNPANST LVNMDSGYSQ 300 KDIVSHNQSI MTPANRDSEA LPAKPTQEPR SADQPMDRED IAKNHVSKNQ ACDAPMSKVG 360 TSGDRNMEPV VATSSVCSAN SAERTSDEPA RNLKRKSHDT TESEGPSEEA EEESVGARKA 420 GHGRNGSKRS RAAEVHNLSE RRRRDRINEK MRALQELIPN CNKADKASML DEAIEYLKTL 480 QIQVQILSMG AGLYMQPMML PPGMQPIHGA HMPHFSPMGL GMGMGMGFGM NMLNMNNGPK 540 MLPFQGPHYP VPGTGFQGMP GSNLPAFPHP GQVLPMSMQQ APIVPPPISG SFLNMPIGLP 600 ASGVAGPSSA PQLAPPGSKK DANPHGLSSM ASNNVKSSLN PATIQGSNEG NQVCHDQTQA 660 VNAVETASGK DAMHS |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 393 | 397 | KRKSH |
| 2 | 439 | 444 | ERRRRD |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00081 | ChIP-seq | Transfer from AT1G09530 | Download |
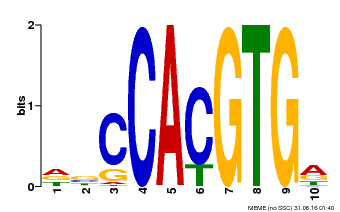
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_019104506.1 | 0.0 | PREDICTED: transcription factor PIF3 isoform X1 | ||||
| Refseq | XP_019104507.1 | 0.0 | PREDICTED: transcription factor PIF3 isoform X1 | ||||
| Refseq | XP_019104508.1 | 0.0 | PREDICTED: transcription factor PIF3 isoform X1 | ||||
| TrEMBL | A0A0K9RD81 | 0.0 | A0A0K9RD81_SPIOL; Uncharacterized protein | ||||
| STRING | XP_010674470.1 | 0.0 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G09530.2 | 6e-37 | phytochrome interacting factor 3 | ||||



