 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Bv4_087770_adcr.t1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Chenopodiaceae; Betoideae; Beta
|
||||||||
| Family | FAR1 | ||||||||
| Protein Properties | Length: 766aa MW: 87577 Da PI: 7.451 | ||||||||
| Description | FAR1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | FAR1 | 83.8 | 2.9e-26 | 79 | 166 | 1 | 91 |
FAR1 1 kfYneYAkevGFsvrkskskkskrngeitkrtfvCskegkreeekkktekerrtraetrtgCkaklkvkkekdgkwevtkleleHnHelap 91
kfY+eYAk++GF++++++s++sk+++ +++++f Cs++g + e++ ++r+ ++t+Cka+++vk++k gkw+++++ +eHnHel p
Bv4_087770_adcr.t1 79 KFYQEYAKSMGFTTSIKNSRRSKKSKDFIDAKFACSRYGATPESDIG---SSRRPGIKKTDCKASMHVKRNKYGKWVIHEFIKEHNHELLP 166
6***************************************9999887...77788899******************************975 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF03101 | 9.5E-24 | 79 | 166 | IPR004330 | FAR1 DNA binding domain |
| Pfam | PF10551 | 7.6E-7 | 243 | 297 | IPR018289 | MULE transposase domain |
| PROSITE profile | PS50966 | 9.31 | 491 | 527 | IPR007527 | Zinc finger, SWIM-type |
| Pfam | PF04434 | 1.6E-5 | 492 | 525 | IPR007527 | Zinc finger, SWIM-type |
| SMART | SM00575 | 5.4E-8 | 502 | 529 | IPR006564 | Zinc finger, PMZ-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010018 | Biological Process | far-red light signaling pathway | ||||
| GO:0042753 | Biological Process | positive regulation of circadian rhythm | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008270 | Molecular Function | zinc ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 766 aa Download sequence Send to blast |
MDLLTKDSHP GVAGSVIDQR GDICHAEGGS RNMADLVNVV HDRDVGTVAS ASSSLLLEGD 60 IDIGPDKDLQ FESHEAAYKF YQEYAKSMGF TTSIKNSRRS KKSKDFIDAK FACSRYGATP 120 ESDIGSSRRP GIKKTDCKAS MHVKRNKYGK WVIHEFIKEH NHELLPALAY YFRIHRNVKM 180 VEKNNIDILH AVSERTKKMY VDMSRQSASF LNDGSLRDAH GSQFDKGNQL VLDEGDAQFL 240 LDYFMCVKKE NPKFFYAVHL NEEQAMGGGA PQVIITDQHP SIKSAIQEIF PDTRHCFHLC 300 NVLEKVPENL AHVTRRHENF HRKFNKCIFK SWSDEQFELR WWKMVTRFDL QEDDWVRSLY 360 EDRKKWVPLY MSGIFLAGMS PTHRSESVNS FFDKYIQRKL TLKEFVKQYG VVLQNRYEEE 420 AIADFDTLHK QPALKSPSPW EKQMSIIYTH AIFKKFQVEV LGVVGCHPKK EGEDGTSITY 480 RVEDCEKAET FFVAWDETKS QVSCLCRSFE YKGFLCRHAM LVLQICGISS IPSQYILRRW 540 TKDAKTRQLM VEGTERLQTR AQRYVSLCKQ AIGLSEEGSL TQDSHSIALR TLVDTLKNCV 600 RMNNIHRSAT ELGDTYCAHD IVQTFQENIS TRTSKKKNVV KKRKVASETD VMLTDAEDNL 660 HHMENLSSDG ISLNGYYGSQ PNVPGLVQLN LMEPPHDSFY VDQESMQGLG QLNSIGPNHE 720 GFFGAQQSVH GLGHLDFRPS SSFSYSLQDE HNLRSTELRG DASRHT |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
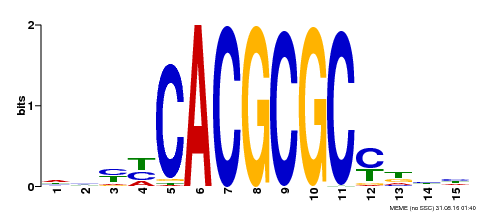
| UniProt | Transcription activator that recognizes and binds to the DNA consensus sequence 5'-CACGCGC-3'. Activates the expression of FHY1 and FHL involved in light responses. Positive regulator of chlorophyll biosynthesis via the activation of HEMB1 gene expression. {ECO:0000269|PubMed:11889039, ECO:0000269|PubMed:12753585, ECO:0000269|PubMed:18033885, ECO:0000269|PubMed:22634759}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00434 | DAP | Transfer from AT4G15090 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Down-regulated after exposure to far-red light. Subject to a negative feedback regulation by PHYA signaling. {ECO:0000269|PubMed:18033885}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010675660.1 | 0.0 | PREDICTED: protein FAR-RED IMPAIRED RESPONSE 1 isoform X1 | ||||
| Refseq | XP_010675661.1 | 0.0 | PREDICTED: protein FAR-RED IMPAIRED RESPONSE 1 isoform X1 | ||||
| Refseq | XP_010675662.1 | 0.0 | PREDICTED: protein FAR-RED IMPAIRED RESPONSE 1 isoform X1 | ||||
| Refseq | XP_010675663.1 | 0.0 | PREDICTED: protein FAR-RED IMPAIRED RESPONSE 1 isoform X1 | ||||
| Refseq | XP_019104685.1 | 0.0 | PREDICTED: protein FAR-RED IMPAIRED RESPONSE 1 isoform X1 | ||||
| Refseq | XP_019104687.1 | 0.0 | PREDICTED: protein FAR-RED IMPAIRED RESPONSE 1 isoform X1 | ||||
| Swissprot | Q9SWG3 | 0.0 | FAR1_ARATH; Protein FAR-RED IMPAIRED RESPONSE 1 | ||||
| TrEMBL | A0A0K9QKJ9 | 0.0 | A0A0K9QKJ9_SPIOL; Uncharacterized protein | ||||
| STRING | XP_010675660.1 | 0.0 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G15090.1 | 0.0 | FAR1 family protein | ||||



