 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Bv6_128480_kgcj.t1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Chenopodiaceae; Betoideae; Beta
|
||||||||
| Family | AP2 | ||||||||
| Protein Properties | Length: 696aa MW: 77721.2 Da PI: 7.0471 | ||||||||
| Description | AP2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 52.2 | 1.5e-16 | 361 | 420 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrd.pse.ng..kr.krfslgkfgtaeeAakaaiaarkkleg 55
s+y+GV++++++gr++A+++d ++ g ++ ++++lg ++t+e+Aa+a++ a++k++g
Bv6_128480_kgcj.t1 361 SQYRGVTRHRWTGRYEAHLWDnSCKkEGqsRKgRQVYLGGYDTEEKAARAYDMAALKYWG 420
78*******************988886678446*************************98 PP
| |||||||
| 2 | AP2 | 46.3 | 1e-14 | 463 | 514 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
s y+GV+++++ grW A+I + +k +lg+f t eeAa+a++ a+ k++g
Bv6_128480_kgcj.t1 463 SIYRGVTRHHQHGRWQARIGRVAG---NKDLYLGTFSTQEEAAEAYDIAAIKFRG 514
57****************988532...5************************997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00018 | 5.87E-23 | 361 | 428 | No hit | No description |
| SuperFamily | SSF54171 | 1.7E-17 | 361 | 430 | IPR016177 | DNA-binding domain |
| Pfam | PF00847 | 8.3E-13 | 361 | 420 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 4.0E-17 | 362 | 429 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 19.849 | 362 | 428 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 1.4E-30 | 362 | 434 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 1.0E-5 | 363 | 374 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 2.55E-17 | 463 | 522 | IPR016177 | DNA-binding domain |
| CDD | cd00018 | 3.34E-25 | 463 | 524 | No hit | No description |
| Pfam | PF00847 | 1.5E-9 | 463 | 514 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 6.5E-18 | 464 | 522 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 19.019 | 464 | 522 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 4.8E-34 | 464 | 528 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 1.0E-5 | 504 | 524 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0007276 | Biological Process | gamete generation | ||||
| GO:0010492 | Biological Process | maintenance of shoot apical meristem identity | ||||
| GO:0042127 | Biological Process | regulation of cell proliferation | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 696 aa Download sequence Send to blast |
MKSMNNDNDG ISNNNNGWLL GFSLSPQINN LNQQQHQQHQ QHQQQQQQHQ QHQQHVSQES 60 HHHQTPTTTS SVSTSLATLN SFFPSHHNFH QQNNNNIYNH YYGGSATTSV EETEVVHNHD 120 SNHNSSFLYS HPFSLMPLKS DGSLCLPQGM VQAIGSCSTT TSTTTSSATP KLEDFFGDDN 180 HQHQHHHHYN HHHDHHQPQQ QHEVVHVQQY PFYPQLYAGA LQGKEDQVIG DHHQLSSTMN 240 NETDISAGLR NWVQRSYIGC IGSDNGDQIN GGDQSGVTMN TGGGSIGYAD LQSLSLSMSP 300 GSQSSCVTAS HQITPSSTTV IGSESLGIME SSRKRGGERM DQTTKPIVHR KSLDTFGQRT 360 SQYRGVTRHR WTGRYEAHLW DNSCKKEGQS RKGRQVYLGG YDTEEKAARA YDMAALKYWG 420 PSTHINFPLE NYQQQLEDMK NMTRQEFVAH LRRKSSGFSR GASIYRGVTR HHQHGRWQAR 480 IGRVAGNKDL YLGTFSTQEE AAEAYDIAAI KFRGVNAVTN FDIVRYDVDR IMASSTLLAS 540 ELARRTKALP PPPPPPPQLP AKNDEEQQKP DWKIVLYHSS QQDQIDQASM GMDLAQNHDT 600 TMLGSTAHQE IGGAHFSNTS SLVTSLSSSR EDSPEKNNTT TNSSLPLSFT MPPSATKLFA 660 NPSPNIGTWI NQPVELRTPS ISLPQMPVFA AWADLH |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that recognizes and binds to the DNA consensus sequence 5'-CAC[AG]N[AT]TNCCNANG-3'. Required for the initiation and growth of ovules integumenta, and for the development of female gametophyte. Plays a critical role in the development of gynoecium marginal tissues (e.g. stigma, style and septa), and in the fusion of carpels and of medial ridges leading to ovule primordia. Also involved in organs initiation and development, including floral organs. Maintains the meristematic competence of cells and consequently sustains expression of cell cycle regulators during organogenesis, thus controlling the final size of each organ by controlling their cell number. Regulates INO autoinduction and expression pattern. As ANT promotes petal cell identity and mediates down-regulation of AG in flower whorl 2, it functions as a class A homeotic gene. {ECO:0000269|PubMed:10528263, ECO:0000269|PubMed:10639184, ECO:0000269|PubMed:10948255, ECO:0000269|PubMed:11041883, ECO:0000269|PubMed:12183381, ECO:0000269|PubMed:12271029, ECO:0000269|PubMed:12655002, ECO:0000269|PubMed:8742706, ECO:0000269|PubMed:8742707, ECO:0000269|PubMed:9001406, ECO:0000269|PubMed:9093862, ECO:0000269|PubMed:9118807}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00088 | SELEX | Transfer from AT4G37750 | Download |
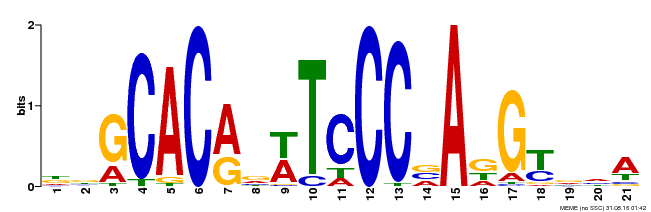
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010679674.1 | 0.0 | PREDICTED: AP2-like ethylene-responsive transcription factor ANT | ||||
| Swissprot | Q38914 | 1e-144 | ANT_ARATH; AP2-like ethylene-responsive transcription factor ANT | ||||
| TrEMBL | A0A0K9QLK8 | 0.0 | A0A0K9QLK8_SPIOL; Uncharacterized protein | ||||
| STRING | XP_010679674.1 | 0.0 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G37750.1 | 1e-126 | AP2 family protein | ||||



