 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Bv6_134560_dzpj.t1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Chenopodiaceae; Betoideae; Beta
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 331aa MW: 36687.2 Da PI: 6.5226 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 103.7 | 1.1e-32 | 197 | 252 | 1 | 56 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRla 56
k r++W+peLH+rFv+a+eqLGGs++AtPk+i+elm+v+gLt+++vkSHLQkYRl+
Bv6_134560_dzpj.t1 197 KRRRCWSPELHRRFVDALEQLGGSQVATPKQIRELMQVDGLTNDEVKSHLQKYRLH 252
689***************************************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 15.396 | 194 | 254 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.64E-18 | 194 | 255 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 8.3E-30 | 195 | 255 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 1.2E-26 | 197 | 252 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 3.9E-8 | 199 | 250 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010092 | Biological Process | specification of organ identity | ||||
| GO:0010629 | Biological Process | negative regulation of gene expression | ||||
| GO:0048449 | Biological Process | floral organ formation | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005829 | Cellular Component | cytosol | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 331 aa Download sequence Send to blast |
MASDFPILSL DLRPPTSSPA ISSSFSDVLT RILVTGIDNS GELSKLSDFV VGLEDELKKI 60 QAFQRELPLS FLLLNDAISK LKEAVKKDEE KRWDGKFGGH IGVKLGIDHE FEKKTWMSSA 120 QLWSPTSREG NKNEERGVNL SFQEATDDVS GLSLVPPGVV AVEKPGTGGI YSGSVFSTGE 180 NQRDKPEMKS AYQTARKRRR CWSPELHRRF VDALEQLGGS QVATPKQIRE LMQVDGLTND 240 EVKSHLQKYR LHVRKLPAGS KSINTLLSTG VDVSCGDQLE LGCSMDLEHS SSPKGPFHLE 300 RSRKCNSSSG EEDPEEEKSD GQSWKARHYQ V |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4r_A | 5e-14 | 197 | 250 | 1 | 54 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_B | 5e-14 | 197 | 250 | 1 | 54 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_C | 5e-14 | 197 | 250 | 1 | 54 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_D | 5e-14 | 197 | 250 | 1 | 54 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 195 | 199 | RKRRR |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00475 | DAP | Transfer from AT4G37180 | Download |
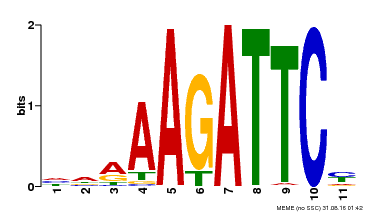
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010680444.1 | 0.0 | PREDICTED: myb family transcription factor EFM | ||||
| TrEMBL | A0A0K9R761 | 1e-132 | A0A0K9R761_SPIOL; Uncharacterized protein | ||||
| STRING | XP_010680444.1 | 0.0 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G37180.1 | 5e-49 | G2-like family protein | ||||



