 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Bv6_155740_wuuo.t1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Chenopodiaceae; Betoideae; Beta
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 402aa MW: 44169.2 Da PI: 6.7874 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 103.5 | 1.3e-32 | 231 | 286 | 1 | 56 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRla 56
k r++W+peLH+rFv+a++qLGGs++AtPk+i++lmkv+gLt+++vkSHLQkYRl+
Bv6_155740_wuuo.t1 231 KSRRCWSPELHRRFVHALQQLGGSHVATPKQIRDLMKVDGLTNDEVKSHLQKYRLH 286
68****************************************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 5.0E-29 | 228 | 289 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 14.267 | 228 | 288 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.08E-17 | 229 | 289 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 2.1E-26 | 231 | 286 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 2.7E-7 | 233 | 284 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 402 aa Download sequence Send to blast |
MMDFRDNMEK CQEYINGLEE EKRKIQVFQR ELPLCLDLVT QAIEACKQQL CGSGTNTTTE 60 CLNGHSECSE QTTSDGPVLE EFIPLKRSNS PDDDDYELES DKSQINSDLN NNHENSGKKS 120 DWLRSVQLWN QTPDPPTTQE DHQPKKVTVV EVKRNGNGAF HPFQKENNCN GNEKHNNNNN 180 NNDNNNKQDA STPAASACSS AQEVDMGPSS GGSGGGSASK KEEKGGQSQR KSRRCWSPEL 240 HRRFVHALQQ LGGSHVATPK QIRDLMKVDG LTNDEVKSHL QKYRLHTRRP SSGVHNNANQ 300 QQAPQFVVVG GIWMPPQEYA AVTATTTSGD KTNVSPPNGI YAPIATVAAP SLMHQTSTHR 360 IQPKQQSSQS HCEERASHSE GGARSNSPAT SSSTHTAASH AH |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4k_A | 5e-14 | 231 | 284 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4k_B | 5e-14 | 231 | 284 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_A | 4e-14 | 231 | 284 | 1 | 54 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_B | 4e-14 | 231 | 284 | 1 | 54 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_C | 4e-14 | 231 | 284 | 1 | 54 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_D | 4e-14 | 231 | 284 | 1 | 54 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_A | 5e-14 | 231 | 284 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_C | 5e-14 | 231 | 284 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_D | 5e-14 | 231 | 284 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_F | 5e-14 | 231 | 284 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_H | 5e-14 | 231 | 284 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_J | 5e-14 | 231 | 284 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor involved in phosphate signaling in roots. {ECO:0000250|UniProtKB:Q9FX67}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00160 | DAP | Transfer from AT1G25550 | Download |
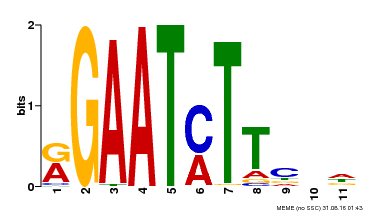
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010695844.1 | 0.0 | PREDICTED: myb family transcription factor EFM | ||||
| Swissprot | Q9FPE8 | 1e-103 | HHO3_ARATH; Transcription factor HHO3 | ||||
| TrEMBL | A0A0J8B7X9 | 0.0 | A0A0J8B7X9_BETVU; Uncharacterized protein | ||||
| STRING | XP_010695844.1 | 0.0 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G25550.1 | 1e-97 | G2-like family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



