 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Bv7_171990_sxrm.t1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Chenopodiaceae; Betoideae; Beta
|
||||||||
| Family | C3H | ||||||||
| Protein Properties | Length: 644aa MW: 70433.6 Da PI: 6.2113 | ||||||||
| Description | C3H family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-CCCH | 21.3 | 4.8e-07 | 279 | 299 | 5 | 26 |
--SGGGGTS--TTTTT-SS-SS CS
zf-CCCH 5 lCrffartGtCkyGdrCkFaHg 26
+C+ f++ G C+ Gd C +aHg
Bv7_171990_sxrm.t1 279 PCPEFRK-GSCPKGDVCEYAHG 299
8******.*************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF48403 | 2.95E-16 | 30 | 154 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 2.9E-17 | 30 | 176 | IPR020683 | Ankyrin repeat-containing domain |
| Pfam | PF12796 | 2.5E-10 | 70 | 146 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 17.29 | 77 | 178 | IPR020683 | Ankyrin repeat-containing domain |
| SMART | SM00248 | 8.5E-4 | 77 | 107 | IPR002110 | Ankyrin repeat |
| CDD | cd00204 | 1.06E-14 | 79 | 157 | No hit | No description |
| PROSITE profile | PS50088 | 12.369 | 112 | 147 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 0.0026 | 112 | 144 | IPR002110 | Ankyrin repeat |
| SMART | SM00356 | 9.4E-5 | 274 | 300 | IPR000571 | Zinc finger, CCCH-type |
| Gene3D | G3DSA:4.10.1000.10 | 5.3E-4 | 278 | 301 | IPR000571 | Zinc finger, CCCH-type |
| PROSITE profile | PS50103 | 12.389 | 279 | 301 | IPR000571 | Zinc finger, CCCH-type |
| Gene3D | G3DSA:4.10.1000.10 | 4.2E-4 | 307 | 337 | IPR000571 | Zinc finger, CCCH-type |
| SMART | SM00356 | 52 | 309 | 332 | IPR000571 | Zinc finger, CCCH-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 644 aa Download sequence Send to blast |
MCSGSKRKVV LAEEVMEGEV FKIDSVEEQQ NSSVLLELSA TNDLSSFKFA VEKEGYDVDE 60 EGLWYGRRVG SKKMGFEERT PLMVAAMFGS KDILNYILQT GRVDVNRASG SDGATALHCA 120 AAGGSLYSHE VIKLLVESSA DVNRVDANGN RPCDVVAPSF SFSSKLKRRT LDVLLNKGDQ 180 GEEGGDQIEE GQQQQQHEGL SPKSSSDGSG SERKEYPIDP SLPDIKNGIY GTDEFRMYTF 240 KVKPCSRAYS HDWTECPFVH PGENARRRDP RKYHYSCVPC PEFRKGSCPK GDVCEYAHGI 300 FECWLHPAQY RTRLCKDETS CTRRVCFFAH KQEELRPLYA STGSAVPSPR SYSQNGPSFD 360 MGSLSPLALG SPSMLMPSTS TPPMTPTGAS SPIGGASAWQ NHLNITPPPA LQLPGSRLKA 420 ALSARDMDFE AGFRRQQLLD ELAGLSLSSP SSWNGELGRL GGLSLSSPTG VQMRQNMNQQ 480 LLSSYSSGLP SSPVRGSSFG LEQSGSPVSN PRLAAFANRS QSFIDRNTVN RLSNGGGMPY 540 HSSDWGSPDG KVDWGVQGDE LNKLRKSASF AFRNNGSNSG PMRNAVNEPD VSWVHSLVKD 600 APPATSAQLG FEEQLKEQYH LNSGMDRIPP WVEQLYMDQE PMVA |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00566 | DAP | Transfer from AT5G58620 | Download |
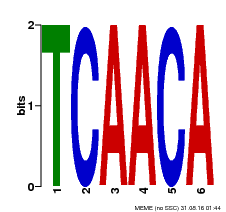
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010685536.1 | 0.0 | PREDICTED: zinc finger CCCH domain-containing protein 66 | ||||
| Swissprot | Q9LUZ4 | 0.0 | C3H66_ARATH; Zinc finger CCCH domain-containing protein 66 | ||||
| TrEMBL | A0A0K9QQB9 | 0.0 | A0A0K9QQB9_SPIOL; Uncharacterized protein | ||||
| STRING | XP_010685536.1 | 0.0 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G58620.1 | 0.0 | C3H family protein | ||||



