 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Bv7_179610_tysn.t1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Chenopodiaceae; Betoideae; Beta
|
||||||||
| Family | AP2 | ||||||||
| Protein Properties | Length: 431aa MW: 47779.7 Da PI: 6.6562 | ||||||||
| Description | AP2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 53.5 | 5.9e-17 | 120 | 169 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkr.krfslgkfgtaeeAakaaiaarkkleg 55
s+y+GV++++++grW+++I+d + k+++lg f+ta Aa+a+++a+ k++g
Bv7_179610_tysn.t1 120 SQYRGVTFYRRTGRWESHIWD------NgKQVYLGGFDTAHAAARAYDRAAIKFRG 169
78*******************......44************************997 PP
| |||||||
| 2 | AP2 | 41.9 | 2.5e-13 | 212 | 262 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpseng...krkrfslgkfgtaeeAakaaiaarkkleg 55
s+y+GV+ +k grW+A+ g +k+++lg ++++ eAa+a+++a+++ +g
Bv7_179610_tysn.t1 212 SKYRGVTLHK-CGRWEAR---M---GqllGKKYIYLGLYDSEIEAARAYDKAALQCNG 262
89********.7******...5...334336**********99**********98765 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF00847 | 4.4E-10 | 120 | 169 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 1.05E-16 | 120 | 179 | IPR016177 | DNA-binding domain |
| CDD | cd00018 | 6.22E-12 | 121 | 179 | No hit | No description |
| PROSITE profile | PS51032 | 18.281 | 121 | 177 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 1.3E-17 | 121 | 177 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 2.4E-32 | 121 | 183 | IPR001471 | AP2/ERF domain |
| Pfam | PF00847 | 2.3E-9 | 212 | 262 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 1.70E-24 | 212 | 272 | No hit | No description |
| SuperFamily | SSF54171 | 1.63E-16 | 212 | 272 | IPR016177 | DNA-binding domain |
| PROSITE profile | PS51032 | 15.172 | 213 | 270 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 2.0E-15 | 213 | 270 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 1.1E-28 | 213 | 276 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 5.2E-6 | 214 | 225 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 5.2E-6 | 252 | 272 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 431 aa Download sequence Send to blast |
MLDLNLNAFS PDSTSENLLP ETSFTSNNSS SLDIDNNADS LSSNSTTPYN FDILNNGASI 60 THDHHHPIIN RGEASSWAGL VTKDFFPGPA QPIRIDCGPV MIGQAQKMKK SRRGPRSRSS 120 QYRGVTFYRR TGRWESHIWD NGKQVYLGGF DTAHAAARAY DRAAIKFRGV DADINFDVSD 180 YEDDLKQMNN LSKEEFVHLL RRQSTGFSRG SSKYRGVTLH KCGRWEARMG QLLGKKYIYL 240 GLYDSEIEAA RAYDKAALQC NGKEAVTNFD PSIYGDDMIS ETHDEGCQYS LDLNLGIGNP 300 SFADSQQTPH NPGSYSFLHD QDNRLTNIRP KDQNCIQAAT VPPPHLAHAL PSEHHSSWGS 360 ANPIFFPVYQ ERAIEKTSET DPWSNWAWQI QFQGGHGSAM PVPVFSTAAS SGFPSITTAS 420 SAAVGRTHFS N |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor (By similarity). Involved in spikelet transition (Probable). Together with SNB, controls synergistically inflorescence architecture and floral meristem establishment via the regulation of spatio-temporal expression of B- and E-function floral organ identity genes in the lodicules and of spikelet meristem genes (PubMed:22003982). Prevents lemma and palea elongation as well as grain growth (PubMed:28066457). {ECO:0000250|UniProtKB:P47927, ECO:0000269|PubMed:22003982, ECO:0000269|PubMed:28066457, ECO:0000305|PubMed:26631749}. | |||||
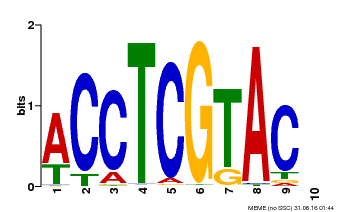
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00615 | PBM | Transfer from AT2G28550 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Target of miR172 microRNA mediated cleavage, particularly during floral organ development. {ECO:0000269|PubMed:22003982, ECO:0000305|PubMed:20017947, ECO:0000305|PubMed:26631749, ECO:0000305|PubMed:28066457}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010696369.1 | 0.0 | PREDICTED: AP2-like ethylene-responsive transcription factor TOE3 | ||||
| Swissprot | Q84TB5 | 1e-101 | AP22_ORYSJ; APETALA2-like protein 2 | ||||
| TrEMBL | A0A0J8B706 | 0.0 | A0A0J8B706_BETVU; Uncharacterized protein | ||||
| STRING | XP_010696369.1 | 0.0 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G28550.3 | 4e-88 | related to AP2.7 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



