 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Bv9_209620_ccoj.t1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Chenopodiaceae; Betoideae; Beta
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 432aa MW: 47449.9 Da PI: 6.6047 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 88.4 | 6.6e-28 | 174 | 228 | 1 | 56 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRla 56
k++++WtpeLH+rFv+aveqL G +kA+P++ilelm++++Lt+++++SHLQkYR++
Bv9_209620_ccoj.t1 174 KTKVDWTPELHRRFVQAVEQL-GVDKAVPSRILELMGIDCLTRHNIASHLQKYRSH 228
6799*****************.********************************86 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 17.937 | 171 | 230 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 8.06E-19 | 172 | 231 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 5.4E-27 | 172 | 231 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 1.6E-26 | 174 | 228 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 4.6E-8 | 177 | 226 | IPR001005 | SANT/Myb domain |
| PROSITE pattern | PS00074 | 0 | 251 | 264 | IPR033524 | Leu/Phe/Val dehydrogenases active site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0007165 | Biological Process | signal transduction | ||||
| GO:0009658 | Biological Process | chloroplast organization | ||||
| GO:0009910 | Biological Process | negative regulation of flower development | ||||
| GO:0010380 | Biological Process | regulation of chlorophyll biosynthetic process | ||||
| GO:0010638 | Biological Process | positive regulation of organelle organization | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:1900056 | Biological Process | negative regulation of leaf senescence | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 432 aa Download sequence Send to blast |
MLAVSPLSTT KDGNNKGDAM EANFTVGSDD FPDFSGANLL DSIDFDELFM GMHDGDMLPD 60 LEMDPELLAE FSISGGEESS EANNSSTMSV DNIISPTNDG HLTLLETNDV ISDVLEKEFM 120 EKHGLMLVKF PEEEIQSKKD VKSTKESDKV KKSSSNSKSG ANNNNSNNNH GKRKTKVDWT 180 PELHRRFVQA VEQLGVDKAV PSRILELMGI DCLTRHNIAS HLQKYRSHRK HLLAREAEAA 240 SWTQKRQMYG VAMGGSKGGA NISPWYLPTM GFPPITTMHP PFRPLHVWGH PTMDPQARMP 300 MWPKQVVPSA APLPPGWAPP PPHPPPADPS FWHSSHQRRV ATAPVRGIPP QTMYRPEQRG 360 VPPHPPPAPA PQSGSHPSFE LHPSKESIDA ALGDVLAKPW LPLPIGLKPP SMDCVLVELQ 420 RQGVPKIPPS CA |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1irz_A | 6e-16 | 171 | 226 | 2 | 57 | ARR10-B |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00022 | PBM | Transfer from AT2G20570 | Download |
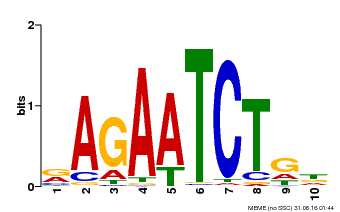
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010689789.1 | 0.0 | PREDICTED: probable transcription factor GLK1 isoform X2 | ||||
| TrEMBL | A0A0K9RD60 | 0.0 | A0A0K9RD60_SPIOL; Uncharacterized protein | ||||
| STRING | XP_010689788.1 | 0.0 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G20570.1 | 2e-71 | GBF's pro-rich region-interacting factor 1 | ||||



