 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Bv9_221790_dgis.t1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Chenopodiaceae; Betoideae; Beta
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 836aa MW: 92002.6 Da PI: 6.4187 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 59.3 | 6.5e-19 | 17 | 75 | 3 | 57 |
--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHC....TS-HHHHHHHHHHHHHHHHC CS
Homeobox 3 kRttftkeqleeLeelFeknrypsaeereeLAkkl....gLterqVkvWFqNrRakekk 57
k ++t+eq+e+Le+l++++++ps +r++L +++ +++ +q+kvWFqNrR +ek+
Bv9_221790_dgis.t1 17 KYVRYTPEQVEALERLYHECPKPSSLRRQQLIRECpilsNIEPKQIKVWFQNRRCREKQ 75
5679*****************************************************97 PP
| |||||||
| 2 | START | 178.4 | 4.3e-56 | 161 | 369 | 2 | 205 |
HHHHHHHHHHHHHHHC-TT-EEEEEXCCTTEEEEEEESSS.SCEEEEEEEECCSCHHHHHHHHHCCCGGCT-TT-SEEEEEEEECTT..EE CS
START 2 laeeaaqelvkkalaeepgWvkssesengdevlqkfeeskvdsgealrasgvvdmvlallveellddkeqWdetlakaetlevissg..ga 90
+aee+++e+++ka+ ++ Wv+++ +++g++++ +++ s++++g a+ra+g+v +++ v+e+l+d++ W ++++++++l+v+ ++ g+
Bv9_221790_dgis.t1 161 IAEETLAEFLSKATGTAVEWVQMPGMKPGPDSIGIVAISHGCTGVAARACGLVGLEPT-RVAEILKDRPSWFRDCRAVDVLNVLPTAngGT 250
7899******************************************************.8888888888****************9999** PP
EEEEEEXXTTXX-SSX.EEEEEEEEEEE.TTS-EEEEEEEEE-TTS--....-TTSEE-EESSEEEEEEEECTCEEEEEEEE-EE--SSXX CS
START 91 lqlmvaelqalsplvp.RdfvfvRyirqlgagdwvivdvSvdseqkppe...sssvvRaellpSgiliepksnghskvtwvehvdlkgrlp 177
++l +++l+a+++l+p Rdf+ +Ry+ +++g++v+ ++S++++q+ p+ +++vRae+lpSg+li+p+++g+s +++v+h+dl+ +++
Bv9_221790_dgis.t1 251 IELLYMQLYAPTTLAPaRDFWLLRYTSVMEDGSLVVSERSLNNTQNGPSmppVQNFVRAEMLPSGYLIRPCEGGGSIIHIVDHMDLEPWSV 341
***********************************************9988899************************************* PP
HHHHHHHHHHHHHHHHHHHHHHTXXXXX CS
START 178 hwllrslvksglaegaktwvatlqrqce 205
+++lr+l++s+++ ++kt++a+l+++++
Bv9_221790_dgis.t1 342 PEVLRPLYESSTVLAQKTTMAALRQLRQ 369
************************9876 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50071 | 15.548 | 12 | 76 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 1.3E-15 | 14 | 80 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 1.25E-16 | 17 | 77 | No hit | No description |
| SuperFamily | SSF46689 | 5.56E-17 | 17 | 80 | IPR009057 | Homeodomain-like |
| Pfam | PF00046 | 1.6E-16 | 18 | 75 | IPR001356 | Homeobox domain |
| Gene3D | G3DSA:1.10.10.60 | 1.0E-18 | 19 | 75 | IPR009057 | Homeodomain-like |
| CDD | cd14686 | 1.73E-6 | 69 | 108 | No hit | No description |
| PROSITE profile | PS50848 | 25.804 | 151 | 366 | IPR002913 | START domain |
| CDD | cd08875 | 6.32E-79 | 155 | 371 | No hit | No description |
| SMART | SM00234 | 4.6E-41 | 160 | 370 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 4.81E-38 | 160 | 372 | No hit | No description |
| Gene3D | G3DSA:3.30.530.20 | 3.5E-24 | 160 | 367 | IPR023393 | START-like domain |
| Pfam | PF01852 | 1.4E-53 | 161 | 369 | IPR002913 | START domain |
| Pfam | PF08670 | 1.7E-51 | 693 | 835 | IPR013978 | MEKHLA |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009965 | Biological Process | leaf morphogenesis | ||||
| GO:0010014 | Biological Process | meristem initiation | ||||
| GO:0010075 | Biological Process | regulation of meristem growth | ||||
| GO:0010087 | Biological Process | phloem or xylem histogenesis | ||||
| GO:0048263 | Biological Process | determination of dorsal identity | ||||
| GO:0080060 | Biological Process | integument development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0008289 | Molecular Function | lipid binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 836 aa Download sequence Send to blast |
MMAMSCKDGK GVMDNGKYVR YTPEQVEALE RLYHECPKPS SLRRQQLIRE CPILSNIEPK 60 QIKVWFQNRR CREKQRKEAS RLQAVNRKLT AMNKLLMEEN DRLQKQVSHL VYENGYFRQQ 120 TQKSGIASKD TSCESVVTSG QHHLTPQHPP RDASPAGLLS IAEETLAEFL SKATGTAVEW 180 VQMPGMKPGP DSIGIVAISH GCTGVAARAC GLVGLEPTRV AEILKDRPSW FRDCRAVDVL 240 NVLPTANGGT IELLYMQLYA PTTLAPARDF WLLRYTSVME DGSLVVSERS LNNTQNGPSM 300 PPVQNFVRAE MLPSGYLIRP CEGGGSIIHI VDHMDLEPWS VPEVLRPLYE SSTVLAQKTT 360 MAALRQLRQI AQEVSQPNVS NWGRRPAALR ALSQRLSRGF NEALNGFTDE GWSMIGNDGI 420 DDVTVLVNSS PDKLMALNLT YANGFPAVSN TVLCAKASML LQNVPPAVLL RFLREHRSEW 480 ADNNIDAYLA AAAKISPCSL PGARIGGFGN QVILPLAHTI EHEELLEVIK LEGVVSCPED 540 AMMPRDMFLL QLCSGMDENA VGTCAELIFA PIDASFADDA PLLPSGFRII PLDSAKEASS 600 PNRTLDLASA LEVGPSANRS SENSGSGNNM RSVMTIAFQF AFESHLQDNV ATMARQYVRS 660 IISSVQRVAL ALSPHLGSQT GLRSPLGNPE AHTLARWICQ SYRCFLGVEL LKSAGEGSDV 720 VLKTLWHHSD AIMCCSLKAL PVFTFANQAG LDMLETTLVA LQDITLEKIL DDHGRKTLCS 780 EFPQILQQGF ACLQGGICLS SMGRPVSYER AVAWKVLNEE ENAHCICFMF MNWSFV |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor involved in the regulation of meristem development to promote lateral organ formation. May regulates procambial and vascular tissue formation or maintenance, and vascular development in inflorescence stems. {ECO:0000269|PubMed:15598805, ECO:0000269|PubMed:15705957, ECO:0000269|PubMed:16617092}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00200 | DAP | Transfer from AT1G52150 | Download |
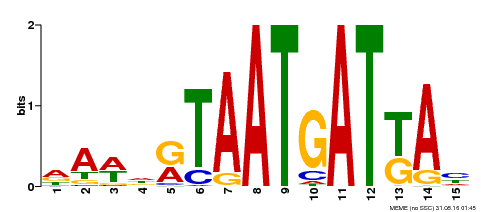
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By auxin. Repressed by miR165 and miR166. {ECO:0000269|PubMed:15773855, ECO:0000269|PubMed:16033795, ECO:0000269|PubMed:16617092, ECO:0000269|PubMed:17237362}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010691423.1 | 0.0 | PREDICTED: homeobox-leucine zipper protein ATHB-15 isoform X1 | ||||
| Swissprot | Q9ZU11 | 0.0 | ATB15_ARATH; Homeobox-leucine zipper protein ATHB-15 | ||||
| TrEMBL | A0A0K9RV39 | 0.0 | A0A0K9RV39_SPIOL; Uncharacterized protein | ||||
| STRING | XP_010691422.1 | 0.0 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G52150.1 | 0.0 | HD-ZIP family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



