 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Bv_008480_ntna.t1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Chenopodiaceae; Betoideae; Beta
|
||||||||
| Family | C3H | ||||||||
| Protein Properties | Length: 426aa MW: 47650.9 Da PI: 6.3723 | ||||||||
| Description | C3H family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-CCCH | 32.1 | 1.9e-10 | 105 | 128 | 4 | 27 |
---SGGGGTS--TTTTT-SS-SSS CS
zf-CCCH 4 elCrffartGtCkyGdrCkFaHgp 27
e C+ff r+GtCk+G +CkF+H+p
Bv_008480_ntna.t1 105 EDCSFFIRNGTCKFGMNCKFNHPP 128
89********************97 PP
| |||||||
| 2 | zf-CCCH | 27.7 | 4.7e-09 | 165 | 187 | 5 | 27 |
--SGGGGTS--TTTTT-SS-SSS CS
zf-CCCH 5 lCrffartGtCkyGdrCkFaHgp 27
+C++f r G Ck+G CkF+H++
Bv_008480_ntna.t1 165 NCKYFDRPGGCKFGKACKFNHSR 187
7********************95 PP
| |||||||
| 3 | zf-CCCH | 39.3 | 1e-12 | 205 | 229 | 2 | 26 |
-S---SGGGGTS--TTTTT-SS-SS CS
zf-CCCH 2 ktelCrffartGtCkyGdrCkFaHg 26
++++C+f++r+G Ck+G++C+F+H+
Bv_008480_ntna.t1 205 GERECPFYMRNGSCKFGASCRFNHP 229
5799********************9 PP
| |||||||
| 4 | zf-CCCH | 30.8 | 5.1e-10 | 332 | 358 | 1 | 27 |
--S---SGGGGTS--TTTTT-SS-SSS CS
zf-CCCH 1 yktelCrffartGtCkyGdrCkFaHgp 27
+++++C+ff++tG Cky + CkF+H++
Bv_008480_ntna.t1 332 PGQPECSFFLKTGDCKYKSACKFNHPK 358
5789*********************96 PP
| |||||||
| 5 | zf-CCCH | 34.2 | 4.4e-11 | 379 | 403 | 2 | 26 |
-S---SGGGGTS--TTTTT-SS-SS CS
zf-CCCH 2 ktelCrffartGtCkyGdrCkFaHg 26
++ C+++ r+G+CkyG+ Ck+ H+
Bv_008480_ntna.t1 379 GENVCSYYNRYGICKYGPACKYDHP 403
6778********************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50103 | 14.784 | 101 | 129 | IPR000571 | Zinc finger, CCCH-type |
| SMART | SM00356 | 3.4E-4 | 101 | 128 | IPR000571 | Zinc finger, CCCH-type |
| SuperFamily | SSF90229 | 1.07E-5 | 103 | 130 | IPR000571 | Zinc finger, CCCH-type |
| Pfam | PF00642 | 3.3E-7 | 105 | 128 | IPR000571 | Zinc finger, CCCH-type |
| Gene3D | G3DSA:4.10.1000.10 | 0.001 | 107 | 127 | IPR000571 | Zinc finger, CCCH-type |
| Gene3D | G3DSA:4.10.1000.10 | 4.3E-5 | 159 | 187 | IPR000571 | Zinc finger, CCCH-type |
| PROSITE profile | PS50103 | 12.859 | 160 | 188 | IPR000571 | Zinc finger, CCCH-type |
| SMART | SM00356 | 0.062 | 161 | 187 | IPR000571 | Zinc finger, CCCH-type |
| Pfam | PF00642 | 6.1E-7 | 165 | 187 | IPR000571 | Zinc finger, CCCH-type |
| SuperFamily | SSF90229 | 7.98E-6 | 203 | 230 | IPR000571 | Zinc finger, CCCH-type |
| SMART | SM00356 | 3.9E-6 | 203 | 230 | IPR000571 | Zinc finger, CCCH-type |
| PROSITE profile | PS50103 | 15.525 | 203 | 231 | IPR000571 | Zinc finger, CCCH-type |
| Pfam | PF00642 | 3.0E-10 | 205 | 229 | IPR000571 | Zinc finger, CCCH-type |
| Gene3D | G3DSA:4.10.1000.10 | 1.4E-4 | 209 | 228 | IPR000571 | Zinc finger, CCCH-type |
| SMART | SM00356 | 2.3E-4 | 331 | 358 | IPR000571 | Zinc finger, CCCH-type |
| PROSITE profile | PS50103 | 14.033 | 331 | 359 | IPR000571 | Zinc finger, CCCH-type |
| Pfam | PF00642 | 2.5E-7 | 333 | 358 | IPR000571 | Zinc finger, CCCH-type |
| SuperFamily | SSF90229 | 7.98E-5 | 335 | 359 | IPR000571 | Zinc finger, CCCH-type |
| SuperFamily | SSF90229 | 7.98E-6 | 375 | 403 | IPR000571 | Zinc finger, CCCH-type |
| PROSITE profile | PS50103 | 14.286 | 377 | 405 | IPR000571 | Zinc finger, CCCH-type |
| SMART | SM00356 | 5.1E-6 | 377 | 404 | IPR000571 | Zinc finger, CCCH-type |
| Pfam | PF00642 | 9.3E-9 | 379 | 403 | IPR000571 | Zinc finger, CCCH-type |
| Gene3D | G3DSA:4.10.1000.10 | 7.2E-4 | 382 | 402 | IPR000571 | Zinc finger, CCCH-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 426 aa Download sequence Send to blast |
MESELESSPI VSPAKQESSS ITSPEPSSPP SDHHKSSDSI NDQLNSLSFE DTKLDNGGED 60 RDHTEERENN EENEESENVS KIEERENEFS RRRSISGFPV RPDAEDCSFF IRNGTCKFGM 120 NCKFNHPPNH RKFQGVKVKA KEECVDSATQ NATENDESLY KEQINCKYFD RPGGCKFGKA 180 CKFNHSRKFN PTLKLNFMGL PIRLGERECP FYMRNGSCKF GASCRFNHPD PTSSGRPDAL 240 SRYGNGGPIF SPDASQSTPA PWSPTALNEN PPYVPLMFSP AQGVPTQPEW NAYQAPVYAP 300 ERSMHMSPIY AMNTRLLERP EHTLGDEFPE RPGQPECSFF LKTGDCKYKS ACKFNHPKSA 360 RSQSSACLLS DLGLPLRPGE NVCSYYNRYG ICKYGPACKY DHPINHAYSP VLGDSMVNNQ 420 AMAAGI |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00582 | DAP | Transfer from AT5G63260 | Download |
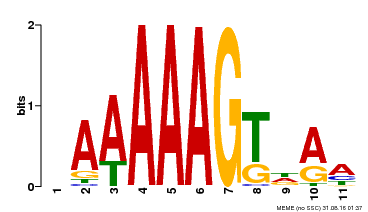
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010667394.1 | 0.0 | PREDICTED: zinc finger CCCH domain-containing protein 67 | ||||
| Swissprot | Q9STM4 | 1e-101 | C3H43_ARATH; Zinc finger CCCH domain-containing protein 43 | ||||
| TrEMBL | A0A0J8B6J5 | 0.0 | A0A0J8B6J5_BETVU; Uncharacterized protein | ||||
| STRING | XP_010667394.1 | 0.0 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G48440.1 | 1e-102 | C3H family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



