 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Bv_008490_cjwm.t1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Chenopodiaceae; Betoideae; Beta
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 1408aa MW: 157640 Da PI: 7.9913 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 13 | 0.00031 | 1317 | 1339 | 3 | 23 |
ET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 3 Cp..dCgksFsrksnLkrHirtH 23
Cp Cgk F ++ +L++H r+H
Bv_008490_cjwm.t1 1317 CPvkGCGKKFFSHKYLVQHRRVH 1339
9999*****************99 PP
| |||||||
| 2 | zf-C2H2 | 12.8 | 0.00036 | 1375 | 1401 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH..T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt..H 23
y+C+ Cg++F+ s++ rH r+ H
Bv_008490_cjwm.t1 1375 YVCNepGCGQTFRFVSDFSRHKRKtgH 1401
99********************99666 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00545 | 1.1E-16 | 25 | 66 | IPR003349 | JmjN domain |
| PROSITE profile | PS51183 | 13.91 | 26 | 67 | IPR003349 | JmjN domain |
| Pfam | PF02375 | 7.2E-14 | 27 | 60 | IPR003349 | JmjN domain |
| SuperFamily | SSF51197 | 3.57E-25 | 162 | 375 | No hit | No description |
| SMART | SM00558 | 6.4E-49 | 192 | 361 | IPR003347 | JmjC domain |
| PROSITE profile | PS51184 | 32.776 | 192 | 361 | IPR003347 | JmjC domain |
| Pfam | PF02373 | 3.8E-37 | 225 | 344 | IPR003347 | JmjC domain |
| Gene3D | G3DSA:3.30.160.60 | 6.0E-4 | 1285 | 1314 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 25 | 1292 | 1314 | IPR015880 | Zinc finger, C2H2-like |
| SMART | SM00355 | 0.0045 | 1315 | 1339 | IPR015880 | Zinc finger, C2H2-like |
| SuperFamily | SSF57667 | 3.17E-6 | 1315 | 1352 | No hit | No description |
| PROSITE profile | PS50157 | 12.445 | 1315 | 1344 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 1.3E-5 | 1316 | 1343 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE pattern | PS00028 | 0 | 1317 | 1339 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 4.3E-8 | 1344 | 1369 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 0.0023 | 1345 | 1369 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 10.72 | 1345 | 1374 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 1347 | 1369 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 1.63E-9 | 1355 | 1399 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 1.4E-9 | 1370 | 1398 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 0.14 | 1375 | 1401 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 11.655 | 1375 | 1406 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 1377 | 1401 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009741 | Biological Process | response to brassinosteroid | ||||
| GO:0009826 | Biological Process | unidimensional cell growth | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0033169 | Biological Process | histone H3-K9 demethylation | ||||
| GO:0035067 | Biological Process | negative regulation of histone acetylation | ||||
| GO:0048366 | Biological Process | leaf development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003676 | Molecular Function | nucleic acid binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1408 aa Download sequence Send to blast |
MGSSSFVTAE HSTPEVFPWL KTLPVAPEYH PTIQEFQDPI AYIFKIEKEA SKFGICKIVP 60 PVPPQSKKSV IANFNRSLES LSDTQKNPKS QPTFTTRQQQ VGFCPRKQRP IHKAVWQSGE 120 YYTLQQFEAK AKNFEKIYLK NRVKKTVSPL EVETLYWKAN GDRPFSVEYA NDMPGSGFVP 180 MKEKKTGEVV ANVGESAWNM RGVARANGSL LRFMKEEIPG VTSPMVYVAM MFSWFAWHVE 240 DHDLHSLNYL HLGASKTWYG CPKDAASAFE EVIRVHGFGE EINPLVTFAT LGEKTTVMSP 300 EVLVNAGVPC CRLVQNAGEF VVTFPRAYHS GFSHGFNCGE ASNIATPEWL RFAKDAAIRR 360 ASINYPPMVS HFQLLYDLAL AICSRVPTGS NAEPRSSRLK DKKKGEGEML VKQMFVHDVI 420 QNNDLLNTLG QGSEVVLLPH NSSEIFVWSN LRVGSKYKVK PGLPFSLYSS EEAIKASDDI 480 MLARDVKQQK GFHSVKTKFG GCTTPHEIQH SETFKGGSSA GDGSDRGLFS CVKCGIWTFA 540 CVAIVQPTES AARYLMSADF SFFNDWIAGS AVSSHGIDAT DGEAYTSDPD SFPGSMEKNA 600 PDSLYDVPVH STDHQARSMD NTSKLKSNTP ETMKVGSDTE MKKESSALGL LAMTYGNSSD 660 SDEDDLLPNC PVVSEDNMSG DGSWGARFHQ DDSASPNFEQ SYDSGVERGP SQVSSRSECE 720 DEDSPQRFEV YGNCGHRRVN GDDNDYESHN CSAKFTEEDT LTSEQHYSPM VDEHDTAKIS 780 CAIDPVSKPN LSFAHRCDED SSRMHVFCLE HALEVEKQLR PIGGVHILLL CHPDYPNVEV 840 EAKKVAEELE MDYVWKDVAF SVATKEDEER IHMALQSEEA TPKNGDWAVK LGINLFYSAI 900 LSRSPLYNKQ MPYNSVIYSA FGCSSPSKSS QEAKVFGKGF GRQKKLVMAG KWCGKVWMSN 960 QVHPLLLHRD LDGEDSVLNP CLKSDEKVGR KSETSYKAPT TDTNRKLGKK RKSMARSNIV 1020 KKMKFEVTEF ADTDPEDSVD DKPEFETRKS VRKPKAQCRP IKRVKHRKED IPDIMSEDSQ 1080 DELEFHQGPL KSARALKQGT KQNLKAKGHQ SVATIDSYSD DDSLDDRSHQ HGLRSCNRKL 1140 KAEGKSTRKV KPQPEEIVDS GESEGSPSDE SEREEHRGNI KSPGYSQRRV PDFKEFEPVT 1200 DDDERDGGPS TRLRRRSVRP TQERRETKAA VKRQVQNSKG KKAPPVTKNA GPARKGPKMR 1260 LPVSRAPAMK GPARKNPSLS SDDANAEEEA EFCCDIEGCT MSFGSKQELA LHKRNICPVK 1320 GCGKKFFSHK YLVQHRRVHM DDRPLKCPWK GCRMTFKWAW ARTEHIRVHT GARPYVCNEP 1380 GCGQTFRFVS DFSRHKRKTG HSAKKTRK |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6ip0_A | 2e-71 | 19 | 419 | 8 | 383 | Transcription factor jumonji (Jmj) family protein |
| 6ip4_A | 2e-71 | 19 | 419 | 8 | 383 | Arabidopsis JMJ13 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00608 | ChIP-seq | Transfer from AT3G48430 | Download |
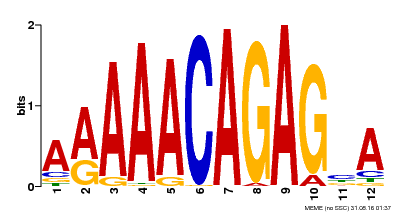
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010667395.1 | 0.0 | PREDICTED: lysine-specific demethylase REF6 isoform X1 | ||||
| TrEMBL | A0A0J8B695 | 0.0 | A0A0J8B695_BETVU; Uncharacterized protein | ||||
| STRING | XP_010667395.1 | 0.0 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G48430.1 | 0.0 | relative of early flowering 6 | ||||



