 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | C.cajan_02909 | ||||||||
| Common Name | KK1_002978 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Cajanus
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 519aa MW: 57531.9 Da PI: 9.4876 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 45.7 | 1.2e-14 | 257 | 307 | 2 | 55 |
HHHHHHHHHHHHHHHHHH.HHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 2 rrahnerErrRRdriNsa.feeLrellPkaskapskKlsKaeiLekAveYIksLq 55
r +h++ E+rRR++iN++ f+ Lrel+P++ ++K +Ka+ L +++eYI+ Lq
C.cajan_02909 257 RSKHSATEQRRRSKINDRqFQMLRELIPHS----DQKRDKASFLLEVIEYIHFLQ 307
889*************9768*********9....9******************99 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.280.10 | 5.8E-22 | 252 | 329 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 1.06E-15 | 253 | 327 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 12.887 | 255 | 306 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 1.68E-13 | 257 | 311 | No hit | No description |
| Pfam | PF00010 | 3.4E-12 | 257 | 307 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 8.4E-11 | 261 | 312 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006351 | Biological Process | transcription, DNA-templated | ||||
| GO:0009742 | Biological Process | brassinosteroid mediated signaling pathway | ||||
| GO:1902448 | Biological Process | positive regulation of shade avoidance | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 519 aa Download sequence Send to blast |
MELPQARPLG TEGRKPTHDF LSLYSNSTAQ QDPRPSSQGS YLKTHDFLQP LERLETKTSA 60 KEEATDEISS AVQKPPTSVE HLLPGGIGTY SISHISYVNN NQRVPKPESS LFVRQATSTD 120 RNDDNSNCSS YTSSGFTLWE ESSGKRGKTG KENNAGEKPT LGESAAKLGQ WTSTERTSQS 180 FSNNRHSSFS SRSSSQTTGQ KNQSFIEMMK SARDSAPDEV LENEETFFLK KEPSNTQREL 240 RVKVDGKSTD QKPNTPRSKH SATEQRRRSK INDRQFQMLR ELIPHSDQKR DKASFLLEVI 300 EYIHFLQEKV HKYEGSFQGW SNEPEKLMPW RNNDKPAENF QPRATDNGSS PSPTLLFASK 360 VDEKNITISP TIPGSTQNLE SGLSTATTSK TMDHQAGIMN NKAFPIPIPS QLNFFTPTQI 420 EKELTIEGGA ISISSVYSKG LLHTLTHALQ SSGVDLSQAS ISVQIELGKQ ANIRPTVPVS 480 MCGAKDGEVH SNNQKMMRSR VASSGKSDQA VKKLKTCRS |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Positive brassinosteroid-signaling protein. Transcription factor that bind specifically to the DNA sequence 5'-CANNTG-3'(E box). Can bind individually to the promoter as a homodimer or synergistically as a heterodimer with BZR2/BES1. Does not itself activate transcription but enhances BZR2/BES1-mediated target gene activation. {ECO:0000269|PubMed:15680330}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00498 | DAP | Transfer from AT5G08130 | Download |
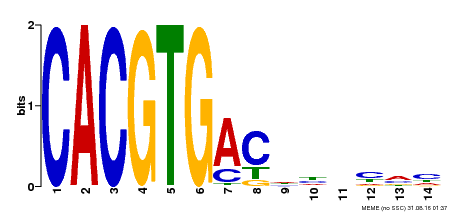
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | C.cajan_02909 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Repressed by cold treatment. {ECO:0000269|PubMed:12679534}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AP015034 | 1e-61 | AP015034.1 Vigna angularis var. angularis DNA, chromosome 1, almost complete sequence, cultivar: Shumari. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020227619.1 | 0.0 | transcription factor BIM1 isoform X2 | ||||
| Swissprot | Q9LEZ3 | 1e-121 | BIM1_ARATH; Transcription factor BIM1 | ||||
| TrEMBL | A0A151SPP9 | 0.0 | A0A151SPP9_CAJCA; Transcription factor BIM1 | ||||
| STRING | GLYMA17G08980.1 | 0.0 | (Glycine max) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF7252 | 31 | 45 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G08130.4 | 1e-118 | bHLH family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



