 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | C.cajan_05797 | ||||||||
| Common Name | KK1_005942 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Cajanus
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 483aa MW: 53961.9 Da PI: 6.574 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 52.6 | 8e-17 | 279 | 325 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
hn ErrRRdriN+++ L++l+P++ K +Ka++Le+A+eY+ksLq
C.cajan_05797 279 VHNLSERRRRDRINEKMRALQQLIPNS-----NKTDKASMLEEAIEYLKSLQ 325
6*************************7.....5******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.280.10 | 4.3E-19 | 272 | 332 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 1.18E-20 | 272 | 338 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 18.463 | 275 | 324 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 4.77E-18 | 278 | 329 | No hit | No description |
| Pfam | PF00010 | 3.0E-14 | 279 | 325 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 7.6E-18 | 281 | 330 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009704 | Biological Process | de-etiolation | ||||
| GO:0010161 | Biological Process | red light signaling pathway | ||||
| GO:0010244 | Biological Process | response to low fluence blue light stimulus by blue low-fluence system | ||||
| GO:0010600 | Biological Process | regulation of auxin biosynthetic process | ||||
| GO:0010928 | Biological Process | regulation of auxin mediated signaling pathway | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 483 aa Download sequence Send to blast |
MNNSIRGWDF ESDTCLNNQR KLVRPDQEFV ELLWQNGQVV MHSQTHKQPV GNSSNLRQVQ 60 KTDQSTLRTS GPYENSSKLD QENAAPWIQY PFEDPSEQEF CSNLLPELPP CEVESYKPIR 120 QMEEGKNANF FASSAPHLTS ISQPLPPNMK PSTSQKTSAN TSQGEAGEYS VITVGSSHCG 180 SNNIPQDQDV SRISSNVHRS DKGKSEMIEP TVTSSSGRSG STGIGRTCSL STRDHGLKRK 240 GTEEEASEEQ SEATELKSAD ENKTSQRTGS SRRNRAAEVH NLSERRRRDR INEKMRALQQ 300 LIPNSNKTDK ASMLEEAIEY LKSLQFQLQV MWMGTGMTPI MFPGIQHYMS QMGLVAPSLP 360 SIHNHMQFPK LAHDQTQMPN QTLMCQNPVL GAFNYQNQMQ NPCLSEQYAR YMGYHLMQSP 420 SQAMNASRYG SEAVQHSQTM IAPTNNSSGP MSGTANVDDA VSGKTSKPLD TFNLFLDFYS 480 QQL |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 283 | 288 | ERRRRD |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00606 | ChIP-seq | Transfer from AT2G43010 | Download |
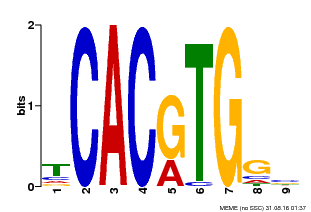
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | C.cajan_05797 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AP015036 | 1e-31 | AP015036.1 Vigna angularis var. angularis DNA, chromosome 3, almost complete sequence, cultivar: Shumari. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020204038.1 | 0.0 | transcription factor PIF4 isoform X1 | ||||
| TrEMBL | A0A151U1W5 | 0.0 | A0A151U1W5_CAJCA; Transcription factor PIF4 | ||||
| STRING | GLYMA02G45150.1 | 0.0 | (Glycine max) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF8146 | 29 | 44 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G43010.2 | 3e-62 | phytochrome interacting factor 4 | ||||



