 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | C.cajan_08718 | ||||||||
| Common Name | KK1_008973 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Cajanus
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 338aa MW: 36957.8 Da PI: 6.8914 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 50.1 | 5.1e-16 | 47 | 96 | 2 | 55 |
HHHHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 2 rrahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
r +h+ E+rRR++iN++f+ Lr+l+P++ ++K + a+ L +++eY+++Lq
C.cajan_08718 47 RSKHSVTEQRRRSKINERFQILRDLIPHS----DQKRDTASFLLEVIEYVQYLQ 96
899*************************9....8*******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF47459 | 1.57E-18 | 43 | 117 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 5.7E-23 | 44 | 117 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 16.044 | 45 | 95 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 4.67E-15 | 47 | 100 | No hit | No description |
| Pfam | PF00010 | 2.3E-13 | 47 | 96 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 4.6E-12 | 51 | 101 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006351 | Biological Process | transcription, DNA-templated | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 338 aa Download sequence Send to blast |
MRTGKGNQEE DEYEEEDFGS SKKQGTSSAS NTNKADGKAI DKASAIRSKH SVTEQRRRSK 60 INERFQILRD LIPHSDQKRD TASFLLEVIE YVQYLQEKVQ KYEGSYQGWG QEPSKLMPWR 120 NSHWRVQSFA GQPTGIKNGL GPVSPFPGKF DESSVNLSPT MLSGTQTTID ADQSRDIVSK 180 TAERQPDLAS KGIPLPVAMP MHANMSVPVR SDGVLAHPLQ GTVSDAQSTE CPTTSEPQNQ 240 QDELTVEGGT ISISSAYSQG LLNNLTQALQ SAGLDLSQAS ISVQINLGKR ANKGLNCGTS 300 SPKNHDNPSS NNQTLAHFRD TGSGEDSDQA QKRMKTYK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Positive brassinosteroid-signaling protein. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00219 | DAP | Transfer from AT1G69010 | Download |
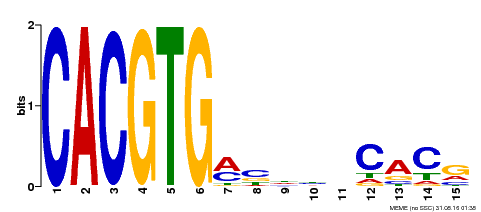
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | C.cajan_08718 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Repressed by heat treatment. {ECO:0000269|PubMed:12679534}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT093538 | 0.0 | BT093538.1 Soybean clone JCVI-FLGm-24O13 unknown mRNA. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020213669.1 | 0.0 | transcription factor BIM2 isoform X1 | ||||
| Refseq | XP_020213670.1 | 0.0 | transcription factor BIM2 isoform X1 | ||||
| Swissprot | Q9CAA4 | 1e-120 | BIM2_ARATH; Transcription factor BIM2 | ||||
| TrEMBL | A0A151TRW5 | 0.0 | A0A151TRW5_CAJCA; Transcription factor BIM2 | ||||
| STRING | GLYMA02G13670.1 | 0.0 | (Glycine max) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1730 | 33 | 82 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G69010.1 | 1e-116 | BES1-interacting Myc-like protein 2 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



