 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | C.cajan_12749 | ||||||||
| Common Name | KK1_013141 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Cajanus
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 371aa MW: 40721.3 Da PI: 6.4546 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 49.6 | 6.8e-16 | 135 | 181 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
hn E+rRR+riN+++ L+ l+P++ K +Ka++L +A+eY+k+Lq
C.cajan_12749 135 VHNLSEKRRRSRINEKMKALQNLIPNS-----NKTDKASMLDEAIEYLKQLQ 181
6*************************7.....5******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF47459 | 1.83E-20 | 128 | 191 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 4.4E-20 | 128 | 191 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 17.86 | 131 | 180 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 5.04E-17 | 134 | 185 | No hit | No description |
| Pfam | PF00010 | 2.3E-13 | 135 | 181 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 8.3E-18 | 137 | 186 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0007623 | Biological Process | circadian rhythm | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0010114 | Biological Process | response to red light | ||||
| GO:0010154 | Biological Process | fruit development | ||||
| GO:0010187 | Biological Process | negative regulation of seed germination | ||||
| GO:0048440 | Biological Process | carpel development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 371 aa Download sequence Send to blast |
MGDMYHFDKN LSSQDEISLF LRQILLRSSP SSSSSSHFMP GSNAPHHNLN AHPSFSPSPL 60 QDGKIPALDS TASFAIKPLH VSAANVSSSP VGVSENENDD YDCESEEGVD ALAEEVPTKP 120 APSRSSSKRS RAAEVHNLSE KRRRSRINEK MKALQNLIPN SNKTDKASML DEAIEYLKQL 180 QLQVQMLSMR NGLSLHPMCY PEGLQLSQMS MELSERNRST PLNMTATLPL HQENPLHYAS 240 NLPNKHAVPN QPSAPYPPYI NNSETSFGLE SRIQPDIKPL QHKGTSEPIR GEDLLHYQQS 300 SAIHSDANTL VGSQVVKEFE SGTTASLSFD TQTCDPKGNN SLHTCIGGRD QSGVVIRNSE 360 ANIVVTSQLS R |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00035 | PBM | Transfer from AT4G36930 | Download |
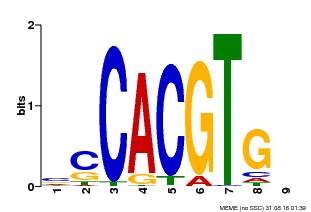
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | C.cajan_12749 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KT031117 | 0.0 | KT031117.1 Glycine max clone HN_CCL_124 bHLH transcription factor (Glyma11g05810.1) mRNA, partial cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020216005.1 | 0.0 | transcription factor SPATULA isoform X1 | ||||
| TrEMBL | A0A151TIE6 | 0.0 | A0A151TIE6_CAJCA; Transcription factor SPATULA | ||||
| STRING | GLYMA11G05810.1 | 0.0 | (Glycine max) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF2276 | 33 | 79 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G36930.1 | 8e-44 | bHLH family protein | ||||



