 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | C.cajan_14927 | ||||||||
| Common Name | KK1_015361 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Cajanus
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 678aa MW: 73688.8 Da PI: 6.2798 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 102.3 | 2.8e-32 | 282 | 339 | 2 | 60 |
--SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS-- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhek 60
dDgynWrKYGqK+vkgse+prsYY+Ct+++C+vkkkvers+ +++++ei+Y+g Hnh+k
C.cajan_14927 282 DDGYNWRKYGQKQVKGSEYPRSYYKCTHPNCQVKKKVERSH-EGHITEIIYKGAHNHPK 339
8****************************************.***************85 PP
| |||||||
| 2 | WRKY | 102.9 | 1.8e-32 | 460 | 518 | 1 | 59 |
---SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 1 ldDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhe 59
ldDgy+WrKYGqK+vkg+++prsYY+Ct agC+v+k+ver+++d k v++tYeg+Hnh+
C.cajan_14927 460 LDDGYRWRKYGQKVVKGNPNPRSYYKCTNAGCTVRKHVERASHDLKSVITTYEGKHNHD 518
59********************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 7.8E-29 | 279 | 340 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 1.01E-25 | 280 | 340 | IPR003657 | WRKY domain |
| SMART | SM00774 | 3.5E-36 | 281 | 339 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 23.502 | 282 | 340 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 7.3E-25 | 282 | 338 | IPR003657 | WRKY domain |
| Gene3D | G3DSA:2.20.25.80 | 8.4E-37 | 445 | 520 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 1.83E-28 | 452 | 520 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 38.32 | 455 | 520 | IPR003657 | WRKY domain |
| SMART | SM00774 | 1.9E-37 | 460 | 519 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 6.8E-25 | 461 | 518 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009555 | Biological Process | pollen development | ||||
| GO:0009942 | Biological Process | longitudinal axis specification | ||||
| GO:0030010 | Biological Process | establishment of cell polarity | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 678 aa Download sequence Send to blast |
MAGIDDNVAL TGDWGLPSPS PRTFFSRMLD EDNVTRSISE HSGNGRTGDL FLGIRDPSET 60 GKGNMKDGAQ DGDSRAQLSE QKSSSRGGLV ERMAARAGFN APRLNTESIR STDLSLNSDI 120 QSPYLTIPPG LSPTTLLDSP VFLANSLAQP SPTTGKFLFM SNSNMRNSEL PSDALEKCKD 180 NGFDMYTSSF AFKPATDSGS SFYHGAGRKI NPTTIPQQSL PGIEVSAQSE NSFQSQTDQM 240 AFDAAGGSTE HSTPIEEQAD EEGDQRVNGD SMSGGVGGAP SDDGYNWRKY GQKQVKGSEY 300 PRSYYKCTHP NCQVKKKVER SHEGHITEII YKGAHNHPKP PPNRRSGIGS VNLQTDMQVD 360 NPEHVEPQNG GDGDLGWTNT QNGTNIDSGE AVDASSTFSN EEDEDDQGTH GSVSLGYDGE 420 GDESESKRRK LESYAELSGA TRAIREPRVV VQTTSEVDIL DDGYRWRKYG QKVVKGNPNP 480 RSYYKCTNAG CTVRKHVERA SHDLKSVITT YEGKHNHDVP AARASSHVNA NASNAVSGQA 540 SGVLQNHVHR PEPSQVHNGI GRLERPSLGS FNLPGRQQLG PSHGFSFGMN QTMLSNFAMS 600 GLGHAQAKLP IMPVHPFLAQ QQQQQQHQHQ HRSANDLGFM MPKGEPNMET IPERGLNLSS 660 GSSVYQEIMS RMPLGPHM |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wj2_A | 1e-36 | 282 | 521 | 8 | 78 | Probable WRKY transcription factor 4 |
| 2lex_A | 1e-36 | 282 | 521 | 8 | 78 | Probable WRKY transcription factor 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor. Regulates WOX8 and WOX9 expression and basal cell division patterns during early embryogenesis. Interacts specifically with the W box (5'-(T)TGAC[CT]-3'), a frequently occurring elicitor-responsive cis-acting element. Required to repolarize the zygote from a transient symmetric state. {ECO:0000269|PubMed:21316593}. | |||||
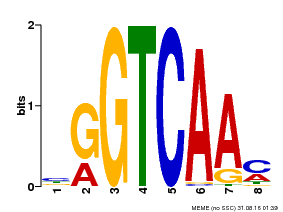
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00071 | PBM | Transfer from AT5G56270 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | C.cajan_14927 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | EU019589 | 1e-145 | EU019589.1 Glycine max WRKY59 (WRKY59) mRNA, partial cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020223534.1 | 0.0 | probable WRKY transcription factor 2 | ||||
| Refseq | XP_020223536.1 | 0.0 | probable WRKY transcription factor 2 | ||||
| Swissprot | Q9FG77 | 0.0 | WRKY2_ARATH; Probable WRKY transcription factor 2 | ||||
| TrEMBL | A0A151SYN7 | 0.0 | A0A151SYN7_CAJCA; Putative WRKY transcription factor 2 | ||||
| STRING | GLYMA04G12830.2 | 0.0 | (Glycine max) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF9524 | 34 | 43 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G56270.1 | 0.0 | WRKY DNA-binding protein 2 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



