 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | C.cajan_20830 | ||||||||
| Common Name | KK1_021452 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Cajanus
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 295aa MW: 31450.8 Da PI: 6.9673 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 35.1 | 3.1e-11 | 11 | 55 | 3 | 45 |
SS-HHHHHHHHHHHHHTTTT...-HHHHHHHHTTTS-HHHHHHHHH CS
Myb_DNA-binding 3 rWTteEdellvdavkqlGgg...tWktIartmgkgRtlkqcksrwq 45
W++e d+ + +a + + + +W++Ia+ ++ g+tl+++k+++
C.cajan_20830 11 EWSKEQDKAFENALAIHAEDasdRWEKIAADVQ-GKTLEEIKHHYE 55
7********************************.**********96 PP
| |||||||
| 2 | Myb_DNA-binding | 43.8 | 5.8e-14 | 122 | 166 | 3 | 47 |
SS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 3 rWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
+WT++E+ l++ + +++G+g+W++I+r + +Rt+ q+ s+ qky
C.cajan_20830 122 AWTEDEHRLFLLGLEKYGKGDWRSISRNFVVTRTPTQVASHAQKY 166
6*******************************************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51293 | 9.906 | 7 | 62 | IPR017884 | SANT domain |
| SMART | SM00717 | 1.8E-8 | 8 | 60 | IPR001005 | SANT/Myb domain |
| SuperFamily | SSF46689 | 9.66E-11 | 10 | 66 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 1.2E-4 | 11 | 57 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 1.33E-8 | 11 | 58 | No hit | No description |
| Pfam | PF00249 | 2.7E-8 | 11 | 55 | IPR001005 | SANT/Myb domain |
| PROSITE profile | PS51294 | 17.61 | 114 | 171 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 8.28E-17 | 117 | 172 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 4.5E-11 | 119 | 169 | IPR001005 | SANT/Myb domain |
| TIGRFAMs | TIGR01557 | 1.5E-17 | 119 | 169 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 7.3E-12 | 122 | 166 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.28E-10 | 122 | 167 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 5.2E-11 | 122 | 165 | IPR009057 | Homeodomain-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 295 aa Download sequence Send to blast |
MTMDEVGSSS EWSKEQDKAF ENALAIHAED ASDRWEKIAA DVQGKTLEEI KHHYELLVED 60 VNQIESGCVP LPSYNSSSEG STSHAGDEGA GKKGGHSWNC NNESNHGTKA SRSDQERRKG 120 IAWTEDEHRL FLLGLEKYGK GDWRSISRNF VVTRTPTQVA SHAQKYFIRL NSMNKDRRRS 180 SIHDITSVNN GDVSAPQGPI TGQTNGSAAN SAGKSTKPAP PTPTAAQGVG IYAAPTIGQP 240 IGGPLVSAVG TPVNLPPPAH MAYSLGATVP GTVVPGAPMN LGPVPYPMPH TSAHR |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2cjj_A | 2e-20 | 7 | 77 | 6 | 76 | RADIALIS |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that coordinates abscisic acid (ABA) biosynthesis and signaling-related genes via binding to the specific promoter motif 5'-(A/T)AACCAT-3'. Represses ABA-mediated salt (e.g. NaCl and KCl) stress tolerance. Regulates leaf shape and promotes vegetative growth. {ECO:0000269|PubMed:26243618}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00500 | DAP | Transfer from AT5G08520 | Download |
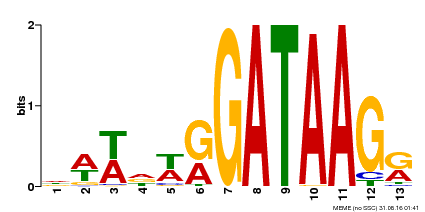
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | C.cajan_20830 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by salicylic acid (SA) and gibberellic acid (GA) (PubMed:16463103). Triggered by dehydration and salt stress (PubMed:26243618). {ECO:0000269|PubMed:16463103, ECO:0000269|PubMed:26243618}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT096693 | 0.0 | BT096693.1 Soybean clone JCVI-FLGm-25B10 unknown mRNA. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020220283.1 | 0.0 | transcription factor SRM1 | ||||
| Refseq | XP_020220361.1 | 0.0 | transcription factor SRM1 | ||||
| Refseq | XP_020220433.1 | 0.0 | transcription factor SRM1 | ||||
| Refseq | XP_029127979.1 | 0.0 | transcription factor SRM1 | ||||
| Swissprot | Q9FNN6 | 1e-127 | SRM1_ARATH; Transcription factor SRM1 | ||||
| TrEMBL | A0A151UD74 | 0.0 | A0A151UD74_CAJCA; Myb-like protein J | ||||
| STRING | GLYMA08G40020.5 | 0.0 | (Glycine max) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF4694 | 33 | 58 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G08520.1 | 1e-128 | MYB family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



