 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | C.cajan_22161 | ||||||||
| Common Name | KK1_022817 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Cajanus
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 222aa MW: 24321.5 Da PI: 5.2457 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 61.6 | 1.8e-19 | 34 | 83 | 2 | 55 |
AP2 2 gykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
+y GVr+++ +gr++AeIrdps+ + r++lg+f+taeeAa a+++a+++++g
C.cajan_22161 34 RYLGVRRRP-WGRYAAEIRDPST---KERHWLGTFDTAEEAALAYDRAARSMRG 83
588******.**********965...5*************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51032 | 21.14 | 34 | 91 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 1.2E-29 | 34 | 90 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 2.48E-21 | 34 | 91 | IPR016177 | DNA-binding domain |
| SMART | SM00380 | 3.7E-34 | 34 | 97 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 7.95E-30 | 34 | 90 | No hit | No description |
| Pfam | PF00847 | 2.2E-12 | 35 | 83 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 7.6E-9 | 35 | 46 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 7.6E-9 | 57 | 73 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009739 | Biological Process | response to gibberellin | ||||
| GO:0010030 | Biological Process | positive regulation of seed germination | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 222 aa Download sequence Send to blast |
MNPYTSSPSS SKSKKKQTTT QQDQQQHETA WGGRYLGVRR RPWGRYAAEI RDPSTKERHW 60 LGTFDTAEEA ALAYDRAARS MRGSRARTNF VYPDTPPGSS VTSIISPDEQ TQTQIQIQIH 120 QAQEELSSLF APNPFPQPDP TPPFSLGGSM GYEMGHGFYG DGVGFSDPSF NVDASGPGYP 180 YSGFESGDYV VHSPLFGAMP PVSDNVATGH EGFDLGSSSY FF |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2gcc_A | 2e-20 | 30 | 90 | 1 | 62 | ATERF1 |
| 3gcc_A | 2e-20 | 30 | 90 | 1 | 62 | ATERF1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Cell division-promoting factor involved in leaf blade differentiation, inflorescence branching, as well as in carpel and silique shape. Promotes the number of xylem cells. Regulates positively the gibberellin signaling pathway leading to germination, hypocotyl elongation, and leaf expansion. Probably acts as a transcriptional activator. Binds to the GCC-box pathogenesis-related promoter element. May be involved in the regulation of gene expression by stress factors and by components of stress signal transduction pathways (By similarity). {ECO:0000250, ECO:0000269|PubMed:11044410, ECO:0000269|PubMed:12472696, ECO:0000269|PubMed:16339853}. | |||||
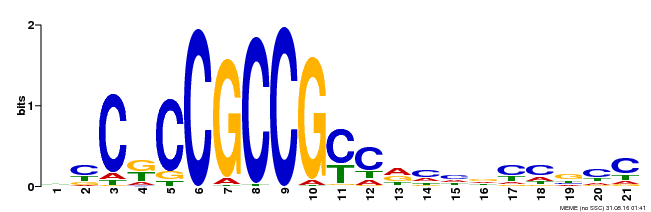
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00509 | DAP | Transfer from AT5G13910 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | C.cajan_22161 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By imbibition. {ECO:0000269|PubMed:16339853}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_003548039.1 | 1e-111 | ethylene-responsive transcription factor LEP | ||||
| Swissprot | Q9M644 | 1e-50 | LEP_ARATH; Ethylene-responsive transcription factor LEP | ||||
| TrEMBL | A0A151T019 | 1e-162 | A0A151T019_CAJCA; Ethylene-responsive transcription factor LEP | ||||
| STRING | GLYMA16G26460.1 | 1e-110 | (Glycine max) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF3904 | 31 | 61 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G13910.1 | 4e-28 | ERF family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



