 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | C.cajan_23192 | ||||||||
| Common Name | KK1_023872 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Cajanus
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 159aa MW: 17726.2 Da PI: 10.8735 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 44.3 | 4.1e-14 | 95 | 139 | 3 | 47 |
SS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 3 rWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
+WT+eE+ +++ + ++lG+g+W+ I+r + ++Rt+ q+ s+ qky
C.cajan_23192 95 PWTEEEHRIFLIGLEKLGKGDWRGISRNFVTTRTPTQVASHAQKY 139
8*******************************************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.60.10 | 1.9E-4 | 3 | 25 | IPR001878 | Zinc finger, CCHC-type |
| PROSITE profile | PS51294 | 19.427 | 88 | 144 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 4.09E-18 | 89 | 144 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 3.3E-18 | 91 | 142 | IPR006447 | Myb domain, plants |
| Gene3D | G3DSA:1.10.10.60 | 7.2E-12 | 92 | 138 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 4.4E-10 | 92 | 142 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.01E-9 | 95 | 140 | No hit | No description |
| Pfam | PF00249 | 2.0E-11 | 95 | 139 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0008270 | Molecular Function | zinc ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 159 aa Download sequence Send to blast |
MGRKCSHCGT IGHNSRTCTS LRGSTSTSFV GLRLFGVQLD TCVAIKKSFS MDSLPSSSFS 60 SSRITTIDDS SDRTSFGYLS DGLLGRPQER KKGVPWTEEE HRIFLIGLEK LGKGDWRGIS 120 RNFVTTRTPT QVASHAQKYF LRLATLNKKK RRSSLFDLV |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 147 | 151 | KKKRR |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription repressor that binds to 5'-TATCCA-3' elements in gene promoters. Contributes to the sugar-repressed transcription of promoters containing SRS or 5'-TATCCA-3' elements. Transcription repressor involved in a cold stress response pathway that confers cold tolerance. Suppresses the DREB1-dependent signaling pathway under prolonged cold stress. DREB1 responds quickly and transiently while MYBS3 responds slowly to cold stress. They may act sequentially and complementarily for adaptation to short- and long-term cold stress (PubMed:20130099). {ECO:0000269|PubMed:12172034, ECO:0000269|PubMed:20130099}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00565 | DAP | Transfer from AT5G56840 | Download |
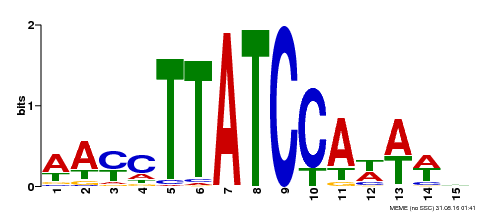
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | C.cajan_23192 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Repressed by sucrose and gibberellic acid (GA) (PubMed:12172034). Induced by cold stress in roots and shoots. Induced by salt stress in shoots. Down-regulated by abscisic aci (ABA) in shoots (PubMed:20130099). {ECO:0000269|PubMed:12172034, ECO:0000269|PubMed:20130099}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KT031194 | 1e-141 | KT031194.1 Glycine max clone HN_CCL_90 MYB/HD-like transcription factor (Glyma08g03330.1) mRNA, partial cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020215967.1 | 1e-114 | transcription factor MYBS3 | ||||
| Swissprot | Q7XC57 | 4e-49 | MYBS3_ORYSJ; Transcription factor MYBS3 | ||||
| TrEMBL | A0A151TKI1 | 1e-111 | A0A151TKI1_CAJCA; Myb-like protein J | ||||
| STRING | GLYMA08G03330.1 | 1e-87 | (Glycine max) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF3213 | 32 | 71 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G56840.1 | 9e-54 | MYB_related family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



