 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | C.cajan_23795 | ||||||||
| Common Name | KK1_024484 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Cajanus
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 194aa MW: 20926.9 Da PI: 9.8223 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 38.8 | 1.7e-12 | 47 | 92 | 5 | 55 |
HHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 5 hnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
h+++Er RR+ri +++ L+el+P+a K +Ka++L + ++Y+k Lq
C.cajan_23795 47 HSIAERLRRERIAERMKALQELVPNA-----NKTDKASMLDEIIDYVKFLQ 92
99***********************8.....6******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00083 | 9.87E-14 | 40 | 96 | No hit | No description |
| SuperFamily | SSF47459 | 1.31E-17 | 40 | 101 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 17.39 | 42 | 91 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 4.8E-17 | 44 | 101 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 7.6E-10 | 47 | 92 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 1.5E-13 | 48 | 97 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 194 aa Download sequence Send to blast |
MPLSFHQTLY NINASPQPPN PHTHTFGPSA PPKHKVRARR GQATDPHSIA ERLRRERIAE 60 RMKALQELVP NANKTDKASM LDEIIDYVKF LQLQVKVLSM SRLGGAAAVA PVVGEMSSEG 120 GKVKGNEASS NDSLAMTEQQ VAKLMEEDMG SAMQYLQGKG LCLMPISLAT AISNATSRTF 180 INAHAPPTSP GPNS |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 51 | 56 | RLRRER |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00659 | PBM | Transfer from Pp3c14_15890 | Download |
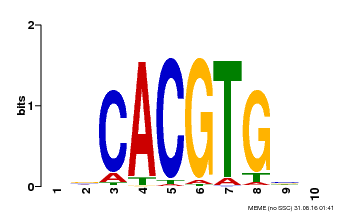
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | C.cajan_23795 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By cold, UV, ethylene (ACC), flagellin, jasmonic acid (JA), and salicylic acid (SA) treatments. {ECO:0000269|PubMed:12679534}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020229201.1 | 1e-141 | transcription factor bHLH82 | ||||
| Swissprot | Q9ZUG9 | 1e-76 | BH066_ARATH; Transcription factor bHLH66 | ||||
| TrEMBL | A0A151SFI3 | 1e-141 | A0A151SFI3_CAJCA; Transcription factor bHLH66 | ||||
| STRING | GLYMA17G08300.2 | 6e-88 | (Glycine max) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF4778 | 33 | 52 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G24260.1 | 9e-62 | LJRHL1-like 1 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



