 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | C.cajan_29441 | ||||||||
| Common Name | KK1_029988 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Cajanus
|
||||||||
| Family | ARF | ||||||||
| Protein Properties | Length: 797aa MW: 88437.6 Da PI: 6.3622 | ||||||||
| Description | ARF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | B3 | 73.8 | 2.1e-23 | 128 | 229 | 1 | 99 |
EEEE-..-HHHHTT-EE--HHH.HTT.......---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-SSSE CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeeh.......ggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkldgrse 89
f+k+lt sd++++g +++p++ ae+ +++++ ++l+ +d++ +W++++i+r++++r++lt+GW+ Fv+a++L +gD+v+F ++++
C.cajan_29441 128 FCKTLTASDTSTHGGFSVPRRAAEKVfppldfsQQPPA--QELIARDLHDVEWKFRHIFRGQPKRHLLTTGWSVFVSAKRLVAGDSVLFI--WNEK 219
99*****************************8544444..48************************************************..8899 PP
E..EEEEE-S CS
B3 90 felvvkvfrk 99
+l+++++r+
C.cajan_29441 220 NQLLLGIRRA 229
999****997 PP
| |||||||
| 2 | Auxin_resp | 120.9 | 9.4e-40 | 254 | 337 | 1 | 83 |
Auxin_resp 1 aahaastksvFevvYnPrastseFvvkvekvekalk.vkvsvGmRfkmafetedsserrlsGtvvgvsdldpvrWpnSkWrsLk 83
aahaa+t+s F+v+YnPras+seFv++++k+ ka++ ++vsvGmRf+m fete+ss rr++Gt++g+sdldpvrWpnS+Wrs+k
C.cajan_29441 254 AAHAAATNSCFTVFYNPRASPSEFVIPLSKYIKAVYhTRVSVGMRFRMLFETEESSVRRYMGTITGISDLDPVRWPNSHWRSVK 337
79*********************************9789******************************************985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101936 | 2.62E-49 | 112 | 257 | IPR015300 | DNA-binding pseudobarrel domain |
| Gene3D | G3DSA:2.40.330.10 | 1.2E-41 | 121 | 243 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 2.32E-21 | 127 | 228 | No hit | No description |
| PROSITE profile | PS50863 | 12.379 | 128 | 230 | IPR003340 | B3 DNA binding domain |
| Pfam | PF02362 | 1.0E-21 | 128 | 229 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 3.4E-24 | 128 | 230 | IPR003340 | B3 DNA binding domain |
| Pfam | PF06507 | 6.5E-35 | 254 | 337 | IPR010525 | Auxin response factor |
| Pfam | PF02309 | 8.1E-8 | 672 | 760 | IPR033389 | AUX/IAA domain |
| SuperFamily | SSF54277 | 7.85E-7 | 674 | 750 | No hit | No description |
| PROSITE profile | PS51745 | 24.155 | 676 | 760 | IPR000270 | PB1 domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009734 | Biological Process | auxin-activated signaling pathway | ||||
| GO:0009908 | Biological Process | flower development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 797 aa Download sequence Send to blast |
MKLSTSGLGQ QGHEGGEKKC LNSELWHACA GPLVSLPTAG TRVVYFPQGH SEQVAATTNR 60 EVDGHIPNYP SLPPQLICQL HNVTMHVDVE TDEVYAQMTL QPLTPQEQKD TFLPMELGIP 120 SKQPSNYFCK TLTASDTSTH GGFSVPRRAA EKVFPPLDFS QQPPAQELIA RDLHDVEWKF 180 RHIFRGQPKR HLLTTGWSVF VSAKRLVAGD SVLFIWNEKN QLLLGIRRAN RPQTVMPSSV 240 LSSDSMHIGL LAAAAHAAAT NSCFTVFYNP RASPSEFVIP LSKYIKAVYH TRVSVGMRFR 300 MLFETEESSV RRYMGTITGI SDLDPVRWPN SHWRSVKVGW DESTAGERQP RVSLWEIEPL 360 TTFPMYPSLF PLRLKRPWHP GTSSFHDGRD EATNGLMWLR GGPGDQALNS LSFQGSGLLP 420 WMQQRMDPTL LGNDHNQQYQ AMFSSGLQNL GTGDLMRQQM MNFQQPFNYL QQSGNPNPPL 480 QLQQPQAIQQ SVSSNNILQP QAQRQHSGLP SPSYSKPDFL DSTMKFSASV SPGQNMLGSL 540 CPEGSGNLLN LSRSGQSMLT EQLPQQSWAP KFTPLQVNAF SNSMPHVQYS GKDTAMVPPH 600 CNSDTQNPII FGVNIDSSGL LLPTTVPRYA TASADTDASA MPLGETGFQG ALYPCVQDSS 660 ELLQSAGQVD PQNQTRTFVK VYKSGSVGRS LDISRFSSYH ELREELAQMF GIEGKLEDPL 720 RSGWQLVFVD RENDVLLLGD DPWESFVNNV WYIKILSPED IQKMGEQAEE SGQRLNSTGA 780 DSHEIVSGLP SIGSLEY |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4ldu_A | 1e-160 | 11 | 358 | 41 | 388 | Auxin response factor 5 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Auxin response factors (ARFs) are transcriptional factors that bind specifically to the DNA sequence 5'-TGTCTC-3' found in the auxin-responsive promoter elements (AuxREs). Seems to act as transcriptional activator. Formation of heterodimers with Aux/IAA proteins may alter their ability to modulate early auxin response genes expression. Regulates both stamen and gynoecium maturation. Promotes jasmonic acid production. Partially redundant with ARF6. Involved in fruit initiation. Acts as an inhibitor to stop further carpel development in the absence of fertilization and the generation of signals required to initiate fruit and seed development. {ECO:0000269|PubMed:12036261, ECO:0000269|PubMed:16107481, ECO:0000269|PubMed:16829592}. | |||||
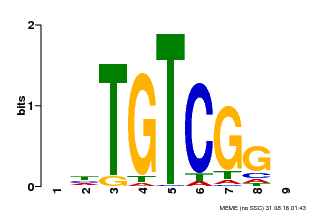
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00033 | PBM | Transfer from AT5G37020 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | C.cajan_29441 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Repressed by miR167. {ECO:0000269|PubMed:17021043}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020234108.1 | 0.0 | auxin response factor 8 isoform X1 | ||||
| Swissprot | Q9FGV1 | 0.0 | ARFH_ARATH; Auxin response factor 8 | ||||
| TrEMBL | A0A151S0V3 | 0.0 | A0A151S0V3_CAJCA; Auxin response factor | ||||
| STRING | GLYMA02G40650.1 | 0.0 | (Glycine max) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF4958 | 28 | 50 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G37020.2 | 0.0 | auxin response factor 8 | ||||



