 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | C.cajan_32294 | ||||||||
| Common Name | KK1_032291 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Cajanus
|
||||||||
| Family | BES1 | ||||||||
| Protein Properties | Length: 1296aa MW: 146270 Da PI: 7.8213 | ||||||||
| Description | BES1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF822 | 135.4 | 6.2e-42 | 663 | 742 | 1 | 81 |
DUF822 1 ggsgrkptwkErEnnkrRERrRRaiaakiyaGLRaqGnyklpkraDnneVlkALcreAGwvvedDGttyrkgskpleeaea 81
+++g+k+++kE+E++k+RER+RRai+++++aGLR++Gn++lp+raD+n+Vl+AL+reAGwvv++DGttyr ++ p++++
C.cajan_32294 663 AAKGKKEREKEKERTKLRERHRRAITSRMLAGLRQYGNFPLPARADMNDVLAALAREAGWVVDADGTTYR-HCPPPSHMVP 742
5899******************************************************************.7887755543 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF57850 | 2.44E-14 | 491 | 546 | No hit | No description |
| Gene3D | G3DSA:3.30.40.10 | 1.1E-12 | 509 | 546 | IPR013083 | Zinc finger, RING/FYVE/PHD-type |
| Pfam | PF13639 | 9.8E-9 | 512 | 546 | IPR001841 | Zinc finger, RING-type |
| CDD | cd00162 | 2.43E-7 | 512 | 549 | No hit | No description |
| PROSITE profile | PS50089 | 9.052 | 513 | 564 | IPR001841 | Zinc finger, RING-type |
| Pfam | PF05687 | 3.2E-44 | 665 | 841 | IPR008540 | BES1/BZR1 plant transcription factor, N-terminal |
| SuperFamily | SSF51445 | 2.46E-156 | 859 | 1293 | IPR017853 | Glycoside hydrolase superfamily |
| Gene3D | G3DSA:3.20.20.80 | 1.5E-169 | 862 | 1292 | IPR013781 | Glycoside hydrolase, catalytic domain |
| Pfam | PF01373 | 7.7E-87 | 884 | 1258 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 1.4E-54 | 899 | 913 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 1.4E-54 | 920 | 938 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 1.4E-54 | 942 | 963 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 1.4E-54 | 1035 | 1057 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 1.4E-54 | 1108 | 1127 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 1.4E-54 | 1142 | 1158 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 1.4E-54 | 1159 | 1170 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 1.4E-54 | 1177 | 1200 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 1.4E-54 | 1215 | 1237 | IPR001554 | Glycoside hydrolase, family 14 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000272 | Biological Process | polysaccharide catabolic process | ||||
| GO:0048831 | Biological Process | regulation of shoot system development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| GO:0008270 | Molecular Function | zinc ion binding | ||||
| GO:0016161 | Molecular Function | beta-amylase activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1296 aa Download sequence Send to blast |
MGSSGSKATS SSSSSGSFRK GRSKGHRGFP SYCLGTTSGS RDIDCDDQVC DQSKVNGDDE 60 TYTSGNEIDS DEGKTESFRK VKSDEVPCVP SNIDLEEWGP TASRTGSSSA HSSSNRSLNT 120 SNGFLSRFSL VPGNISFRLS RTTSLGSSRP CPVSSAGLSI FNNEDELNLH QRPASGLINR 180 NETQHRSNLL NSSFVSQVPI QCHEEGSNNL RPNTPALVSP GNLLSSRTRS SVQDVVRDGD 240 GTREVPDVNF YSPRIHTDTE NFETRHTDRR NGAREPVERN VRFSRTLSVG RLRDRVLRRS 300 TLSDFTICPL QREREVRDAS QDNGRRAGER DTRVSPSGRN AANSSTPRYP QPSTPSSLFG 360 IQDYEVETSR SRETRYQDLL EHRSNFLERR RRIRSQVRAL QRLGSRFENL SGHDRSCILS 420 GQHRNGRCAC RIGSRDTNSN DDTNARASIS RIVMLAEALF EVLDEIHQQS VVLSSRPSVS 480 SIGSVPAPNE VVESLPVKLY TKLHKHQEDP VQCYICLVEY EDGDSMRVLP CHHEFHTTCI 540 DKWLKEIHRP VNQLASNITV LHACDDTLFT FQRKKGKETF QLQCKLRETS VVADPMQDLV 600 DLPCWIPLGL KQKVEGKKRE QKKRNQKMRV NAFHEKPSSV LVLNLPRRPR GFAAATASGA 660 NSAAKGKKER EKEKERTKLR ERHRRAITSR MLAGLRQYGN FPLPARADMN DVLAALAREA 720 GWVVDADGTT YRHCPPPSHM VPFNSTTISR LRFNSLLLIV EYSNLLMVYG GSLAARSVES 780 QLSGGSLRNC SVKETIENQT SVLRIDECLS PASIDSVVIA ERDSKNEKYT NATPINTVDC 840 LEADQLMQDI HSGVHENDFT GTPYVPVYAK LPAGIINKFC QLIDPEGIKQ ELIHIKTLNI 900 DGVVVDCWWG IVEAWSSQKY VWSGYRELFN MIREFKLKLQ VVMAFHECGG NDSSDALISL 960 PQWVLDIGKD NQDIFFTDRE GRRNTECLSW GIDKERVLKG RTGIEVYFDM MRSFRTEFDD 1020 LFAEGLISAV EIGLGASGEL KYPSFSERMG WRYPGIGEFQ CYDKYLQHSL RRAAKLRGHS 1080 FWARGPDNAG HYNSMPHETG FFCERGDYDN YYGRFFLHWY SKTLIDHADN VLSLATLAFE 1140 ETKIIVKVPA VYWWYKTPSH AAELTAGYHN PTNQDGYSPL FEVLRKHAVT MKFVCLGFHL 1200 SSQEANESLI DPEGLSWQVL NSAWDRGLMA VGENALLCYD REGYKRLVEM AKPRNDPDRR 1260 HFSFFVYQQP SLLQANVCLS ELDFFIKCMH GEMTDL |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wdq_A | 1e-121 | 864 | 1254 | 11 | 401 | Beta-amylase |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00543 | DAP | Transfer from AT5G45300 | Download |
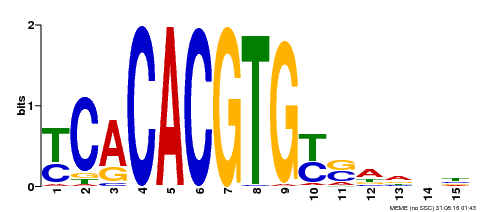
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | C.cajan_32294 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AC173960 | 0.0 | AC173960.15 Glycine max cultivar Williams 82 clone gmw1-45c9, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020236208.1 | 0.0 | beta-amylase 8 | ||||
| TrEMBL | A0A151RUB0 | 0.0 | A0A151RUB0_CAJCA; Beta-amylase | ||||
| STRING | XP_007158095.1 | 0.0 | (Phaseolus vulgaris) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF11902 | 30 | 32 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G45300.1 | 0.0 | beta-amylase 2 | ||||



