 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | C.cajan_34992 | ||||||||
| Common Name | KK1_032750 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Cajanus
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 380aa MW: 41164.5 Da PI: 6.6672 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 54.2 | 3.4e-17 | 38 | 83 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
+g+W++eEd +l ++v ++G ++W++Iar + gR++k+c++rw +
C.cajan_34992 38 KGPWSPEEDAILSRLVSKFGARNWSLIARGIS-GRSGKSCRLRWCNQ 83
79******************************.***********985 PP
| |||||||
| 2 | Myb_DNA-binding | 57.9 | 2.3e-18 | 90 | 134 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
r+++T eEd ++v a++ +G++ W++Iar ++ gRt++ +k++w++
C.cajan_34992 90 RKPFTDEEDRIIVAAHAIHGNK-WAAIARLLP-GRTDNAIKNHWNST 134
789*******************.*********.***********986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 20.375 | 33 | 84 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.11E-30 | 35 | 131 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 6.2E-15 | 37 | 86 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 2.7E-16 | 38 | 83 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 3.5E-26 | 39 | 91 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 2.93E-14 | 40 | 82 | No hit | No description |
| PROSITE profile | PS51294 | 26.069 | 85 | 139 | IPR017930 | Myb domain |
| SMART | SM00717 | 2.2E-17 | 89 | 137 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 2.3E-15 | 90 | 134 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 3.42E-13 | 92 | 135 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 1.1E-23 | 92 | 138 | IPR009057 | Homeodomain-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 380 aa Download sequence Send to blast |
MEPEETAVAV ADDGSGDEGL LGLGGGGSSS GGGRDRVKGP WSPEEDAILS RLVSKFGARN 60 WSLIARGISG RSGKSCRLRW CNQLDPAVKR KPFTDEEDRI IVAAHAIHGN KWAAIARLLP 120 GRTDNAIKNH WNSTLRRRGV ELDKIKLESG NFMEDVSLDK AKASSEDTLS CGDVNSFKSS 180 EGRDVSSVEI MDDKNEDKAQ TEGQLHHEAR DPPTLFRPVA RVSAFSAYHS FDGIQPSASI 240 QRPVPMQGPI LQSTKPDMEF CKMLEGICGD QSVPHQCGHG CCAAQNGKKS KSSLLGPEFI 300 EFSEPPSFPN FELAAIATDI SNLAWLKSGL ENSSVKMMGS TSGRVISNGS QVHIPTLRED 360 WMMGVMKAIV VTIHFPCKCN |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1a5j_A | 2e-41 | 35 | 138 | 4 | 107 | B-MYB |
| Search in ModeBase | ||||||
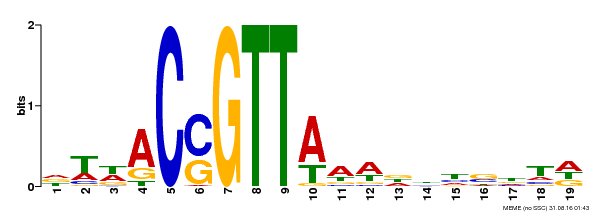
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00337 | DAP | Transfer from AT3G09230 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | C.cajan_34992 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Slightly induced by salicylic acid. {ECO:0000269|PubMed:16463103}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AP015042 | 0.0 | AP015042.1 Vigna angularis var. angularis DNA, chromosome 9, almost complete sequence, cultivar: Shumari. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020236498.1 | 0.0 | transcription factor MYB1 | ||||
| Swissprot | Q42575 | 1e-107 | MYB1_ARATH; Transcription factor MYB1 | ||||
| TrEMBL | A0A151RT46 | 0.0 | A0A151RT46_CAJCA; Transcription factor MYB44 | ||||
| STRING | GLYMA01G26650.1 | 0.0 | (Glycine max) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF3986 | 33 | 62 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G09230.1 | 1e-108 | myb domain protein 1 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



