 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | C.cajan_36291 | ||||||||
| Common Name | KK1_039596 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Cajanus
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 373aa MW: 42452.1 Da PI: 4.9452 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 49.6 | 8.9e-16 | 16 | 63 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
rg+W++ Ed++l+ ++++G ++W++ ++ g+ R++k+c++rw +yl
C.cajan_36291 16 RGPWSPSEDLKLIAFIQKYGHENWRALPKQAGLLRCGKSCRLRWINYL 63
89******************************99************97 PP
| |||||||
| 2 | Myb_DNA-binding | 50.4 | 5.1e-16 | 69 | 114 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
rg++T+ E+e +++++k lG++ W++Ia+ ++ gRt++++k+ w+++l
C.cajan_36291 69 RGNFTPKEEETIIRLHKALGNK-WSKIASSLP-GRTDNEIKNVWNTHL 114
89********************.*********.************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 5.1E-24 | 7 | 66 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 13.87 | 11 | 63 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 2.13E-29 | 14 | 110 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 2.2E-13 | 15 | 65 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.2E-14 | 16 | 63 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.30E-9 | 18 | 63 | No hit | No description |
| PROSITE profile | PS51294 | 24.449 | 64 | 118 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.1E-26 | 67 | 118 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 2.5E-14 | 68 | 116 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.1E-14 | 69 | 114 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.87E-9 | 71 | 114 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:2000652 | Biological Process | regulation of secondary cell wall biogenesis | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 373 aa Download sequence Send to blast |
MGKGRAPCCD KTQVKRGPWS PSEDLKLIAF IQKYGHENWR ALPKQAGLLR CGKSCRLRWI 60 NYLRPDVKRG NFTPKEEETI IRLHKALGNK WSKIASSLPG RTDNEIKNVW NTHLKKRLTA 120 KKCSDSSADE SKPESSITSS SSSSSESFFS NETPNSPKTT TLGNEFNDQA SQVTDKIEQD 180 SDKQVSNELN GIITKDPKES SVSFSSIGSN IVTTSQNVAY KEKQQLASPL SYLGPYDFSS 240 MLEEVDKPNH LFERPWESDF VLWKILRDDS LGSFQSNEVQ LGEFLACQNS ILGEEIVQDV 300 EARKWFHDFE DEFGIVDEIK ESNNDHFLPK NYAVEPEIDH TQTFDFDDIT RPESELDFGY 360 IQLWSSWPQN TSL |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1a5j_A | 1e-26 | 16 | 118 | 7 | 108 | B-MYB |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00249 | DAP | Transfer from AT1G79180 | Download |
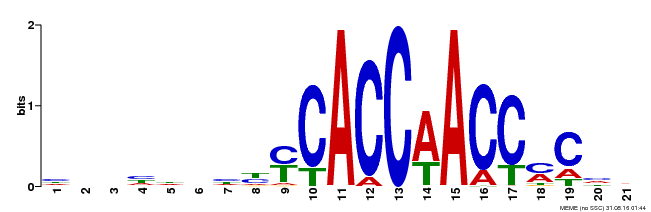
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | C.cajan_36291 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | JN863531 | 2e-75 | JN863531.1 Lotus japonicus R2R3MYB transcription factor (MYB10) mRNA, partial cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020203376.1 | 0.0 | transcription factor MYB58 | ||||
| TrEMBL | A0A151R9E5 | 0.0 | A0A151R9E5_CAJCA; Myb-related protein Zm1 | ||||
| STRING | GLYMA08G17861.1 | 0.0 | (Glycine max) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF10029 | 30 | 42 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G79180.1 | 1e-76 | myb domain protein 63 | ||||



