 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | C.cajan_39946 | ||||||||
| Common Name | KK1_036986 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Cajanus
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 550aa MW: 60251.8 Da PI: 4.9228 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 57.2 | 3.8e-18 | 43 | 90 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+WT Ed++lvd+vk++G g+W+++ ++ g+ R++k+c++rw ++l
C.cajan_39946 43 KGPWTSTEDDILVDYVKKHGEGNWNAVQKHTGLLRCGKSCRLRWANHL 90
79******************************99**********9986 PP
| |||||||
| 2 | Myb_DNA-binding | 51 | 3.3e-16 | 96 | 139 | 1 | 46 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqk 46
+g++T+eE+ l++++++++G++ W++ a++++ gRt++++k++w++
C.cajan_39946 96 KGAFTAEEERLIAELHAKMGNK-WARMAAHLP-GRTDNEIKNYWNT 139
799*******************.*********.***********96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 16.086 | 38 | 90 | IPR017930 | Myb domain |
| SMART | SM00717 | 8.4E-15 | 42 | 92 | IPR001005 | SANT/Myb domain |
| SuperFamily | SSF46689 | 7.86E-30 | 42 | 137 | IPR009057 | Homeodomain-like |
| Pfam | PF00249 | 3.8E-16 | 43 | 90 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.9E-23 | 44 | 97 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 1.27E-11 | 45 | 90 | No hit | No description |
| PROSITE profile | PS51294 | 25.067 | 91 | 145 | IPR017930 | Myb domain |
| SMART | SM00717 | 2.2E-16 | 95 | 143 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 2.9E-15 | 96 | 139 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 4.5E-25 | 98 | 144 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 8.17E-12 | 98 | 139 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009789 | Biological Process | positive regulation of abscisic acid-activated signaling pathway | ||||
| GO:0043068 | Biological Process | positive regulation of programmed cell death | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0045926 | Biological Process | negative regulation of growth | ||||
| GO:0048235 | Biological Process | pollen sperm cell differentiation | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 550 aa Download sequence Send to blast |
MRRLKNDINE DEVLPNDQTG AQLNGAQLND EGSEGSAGVV LKKGPWTSTE DDILVDYVKK 60 HGEGNWNAVQ KHTGLLRCGK SCRLRWANHL RPNLKKGAFT AEEERLIAEL HAKMGNKWAR 120 MAAHLPGRTD NEIKNYWNTR IKRRQRAGLP LYPPEVCLQA LQESQHSQSS GGLNGDNKMH 180 PDFLQKNSYE IQDSIFDSLK DNQGIIPYMP ELSDISVYGN MLKGFDSSQY CSFVPPTSPK 240 HKRLRESTIP FIDSSDMSRN GLYPFDQIRD NNSDEIAQSF GMPAPLDPGP SSHSSICYSH 300 SLSNGNSSTS KPTYEAVKLE LPSLQYPELD LGSWGTSPPP SLLESVDDFI QSPTPISTLE 360 SDCSSPQNSG LLDALLYQAR TLSSSKNHCS DKSSNSSTAT PGDRADSSAL NVHETEWEDY 420 ADPVSPFGAT SILNECPALN ANANSLDKQL PVQTFNGDIL KLESVDEVWT PNGENQTLSL 480 LNITRPDFLL ASDWPDLSSG HGKNLAFTMD ATTTLGLGDD LATDHKHVTA GTSKSSQMWG 540 FGSCATAMIG |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1a5j_A | 1e-31 | 41 | 144 | 5 | 107 | B-MYB |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00490 | DAP | Transfer from AT5G06100 | Download |
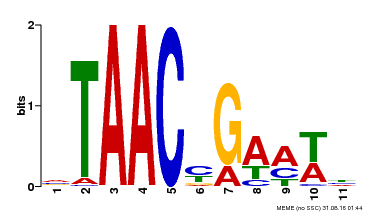
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | C.cajan_39946 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KC525897 | 0.0 | KC525897.1 Glycine max GAMBY1-like family protein (GAMYB1) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020240082.1 | 0.0 | transcription factor MYB33 | ||||
| Refseq | XP_020240083.1 | 0.0 | transcription factor MYB33 | ||||
| Refseq | XP_020240085.1 | 0.0 | transcription factor MYB33 | ||||
| Refseq | XP_029124778.1 | 0.0 | transcription factor MYB33 | ||||
| TrEMBL | A0A151RGG1 | 0.0 | A0A151RGG1_CAJCA; Transcription factor GAMYB | ||||
| STRING | GLYMA13G25716.2 | 0.0 | (Glycine max) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF2047 | 34 | 85 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G06100.3 | 1e-111 | myb domain protein 33 | ||||



