 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | C.cajan_40504 | ||||||||
| Common Name | KK1_038254 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Cajanus
|
||||||||
| Family | RAV | ||||||||
| Protein Properties | Length: 347aa MW: 37705.6 Da PI: 8.9811 | ||||||||
| Description | RAV family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 38.8 | 2.3e-12 | 83 | 131 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
s+ykGV +grW A+I++ ++r++lg+f ++eAa+a++ a+ +++g
C.cajan_40504 83 SKYKGVV-PQLNGRWGAQIYEK-----HQRVWLGTFNEEDEAARAYDVAAVRFRG 131
79****6.7779*********3.....5**********99***********9998 PP
| |||||||
| 2 | B3 | 67.8 | 1.5e-21 | 195 | 261 | 30 | 96 |
..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-SSSEE..EEEE CS
B3 30 keesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkldgrsefelvvkv 96
++++ l +ed g++W+++++y+++s++yvltkGW++Fvk+++Lk+gD+v F+++ +++l++ +
C.cajan_40504 195 AAKGVLLNFEDVGGKVWRFRYSYWNSSQSYVLTKGWSRFVKEKNLKAGDTVCFHRSTGPDKQLYIDW 261
4578899***********************************************8766777777766 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF00847 | 3.0E-7 | 83 | 131 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 4.65E-25 | 83 | 138 | No hit | No description |
| SuperFamily | SSF54171 | 1.31E-16 | 83 | 139 | IPR016177 | DNA-binding domain |
| PROSITE profile | PS51032 | 19.915 | 84 | 139 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 2.4E-27 | 84 | 145 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 3.1E-20 | 84 | 139 | IPR001471 | AP2/ERF domain |
| SMART | SM01019 | 1.8E-7 | 175 | 265 | IPR003340 | B3 DNA binding domain |
| Gene3D | G3DSA:2.40.330.10 | 1.4E-23 | 194 | 264 | IPR015300 | DNA-binding pseudobarrel domain |
| Pfam | PF02362 | 8.7E-19 | 195 | 262 | IPR003340 | B3 DNA binding domain |
| SuperFamily | SSF101936 | 1.44E-18 | 197 | 259 | IPR015300 | DNA-binding pseudobarrel domain |
| PROSITE profile | PS50863 | 11.364 | 201 | 265 | IPR003340 | B3 DNA binding domain |
| CDD | cd10017 | 1.07E-16 | 203 | 250 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009741 | Biological Process | response to brassinosteroid | ||||
| GO:0009910 | Biological Process | negative regulation of flower development | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0048366 | Biological Process | leaf development | ||||
| GO:0048527 | Biological Process | lateral root development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 347 aa Download sequence Send to blast |
MDAISCMDES TTTESLSMSL SPTMSSKKAF VSASPTTSSK KTYVSPPPLS GTLCRVGSGA 60 SAVVDSDSGG GGTEVESRKL PSSKYKGVVP QLNGRWGAQI YEKHQRVWLG TFNEEDEAAR 120 AYDVAAVRFR GKDAVTNFKG LAAAEDDDGE AEFLNSHSKS EIVDMLRKHT YNDELEQSKR 180 SRGVSVNGVN VSATAAKGVL LNFEDVGGKV WRFRYSYWNS SQSYVLTKGW SRFVKEKNLK 240 AGDTVCFHRS TGPDKQLYID WKARNVVVNV NVNVNVNKEV GLFGPVAEPV QMVRLFGVNI 300 LKLPGSECIA NSNNTSGAIP GCCGGKRREM ELFSLECSKK PKIIGAL |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wid_A | 1e-35 | 192 | 264 | 45 | 117 | DNA-binding protein RAV1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Binds specifically to bipartite recognition sequences composed of two unrelated motifs, 5'-CAACA-3' and 5'-CACCTG-3'. May function as negative regulator of plant growth and development. {ECO:0000269|PubMed:15040885, ECO:0000269|PubMed:9862967}. | |||||
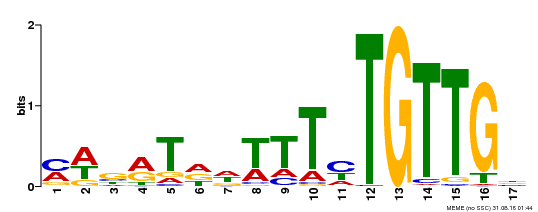
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00139 | DAP | Transfer from AT1G13260 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | C.cajan_40504 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Down-regulated by brassinosteroid and zeatin. {ECO:0000269|PubMed:15040885}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AP015043 | 5e-82 | AP015043.1 Vigna angularis var. angularis DNA, chromosome 10, almost complete sequence, cultivar: Shumari. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020202294.1 | 0.0 | AP2/ERF and B3 domain-containing transcription repressor TEM1 | ||||
| Swissprot | Q9ZWM9 | 1e-108 | RAV1_ARATH; AP2/ERF and B3 domain-containing transcription factor RAV1 | ||||
| TrEMBL | A0A151RDE0 | 0.0 | A0A151RDE0_CAJCA; AP2/ERF and B3 domain-containing transcription factor RAV1 | ||||
| STRING | GLYMA01G22260.1 | 1e-175 | (Glycine max) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF2920 | 32 | 74 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G25730.1 | 3e-82 | ethylene response DNA binding factor 3 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



