 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | C.cajan_44547 | ||||||||
| Common Name | KK1_039032 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Cajanus
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 460aa MW: 49143.8 Da PI: 8.6984 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 48.9 | 1.2e-15 | 277 | 322 | 5 | 55 |
HHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 5 hnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
hn+ Er+RRd+iN+++ +L++l+P++ K +Ka++L +++eY+k+Lq
C.cajan_44547 277 HNQSERKRRDKINQRMKTLQKLVPNS-----SKTDKASMLDEVIEYLKQLQ 322
9************************8.....7******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF47459 | 1.01E-18 | 270 | 334 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 17.815 | 272 | 321 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 3.1E-13 | 277 | 322 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 4.6E-18 | 277 | 330 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 2.63E-17 | 277 | 326 | No hit | No description |
| SMART | SM00353 | 2.2E-18 | 278 | 327 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009567 | Biological Process | double fertilization forming a zygote and endosperm | ||||
| GO:0009506 | Cellular Component | plasmodesma | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 460 aa Download sequence Send to blast |
MSQCVPSWDV DDNPPPPRLS LRSNSNSTAP DVPMLDYEVA ELTWENGQLS MHGLGLPRVP 60 VKPPTSAANK YTWEKPRASG TLESIVNQAT SLPQRGKPSL NGDGVGVCGG FVVPWFDPHA 120 HGGGGGATAN TVGMDALVPC SKREARKQGV ESVPGGTCMV GCSTRVGSCC GNQGAKGHEM 180 SGRDQSVSGS ATFGRDSKHV TLDTCDREFG VAFTSTSINS LDNTSSAKHC TKTTTVDDHD 240 SVSHSKPVGE DGDKEKKKRA NGKSSVSTKR SRAAAIHNQS ERKRRDKINQ RMKTLQKLVP 300 NSSKTDKASM LDEVIEYLKQ LQAQVQMMNR INMSSMMLPL TMQQQLQMSM MSPMGMGLGM 360 GMGMGMGMGM DMNSMNRANI PGIPPVLHPS AFMPMAASWD AAAVAVGGSD RLQGTPATVM 420 PDPLSTFFGC QSQPMNMDAY SRLAAMYQQL HQPPASGSKN |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 280 | 285 | ERKRRD |
| 2 | 281 | 286 | RKRRDK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Required during the fertilization of ovules by pollen. {ECO:0000269|PubMed:15634699}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00034 | PBM | Transfer from AT4G00050 | Download |
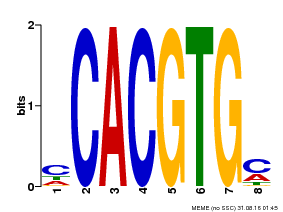
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | C.cajan_44547 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By cold treatment. {ECO:0000269|PubMed:12679534}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AC235052 | 1e-157 | AC235052.1 Glycine max strain Williams 82 clone GM_WBb0119P14, complete sequence. | |||
| GenBank | AC235252 | 1e-157 | AC235252.1 Glycine max strain Williams 82 clone GM_WBb0036F08, complete sequence. | |||
| GenBank | AC235273 | 1e-157 | AC235273.1 Glycine max strain Williams 82 clone GM_WBb0046P21, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020202885.1 | 0.0 | transcription factor UNE10 isoform X1 | ||||
| Swissprot | Q8GZ38 | 1e-108 | UNE10_ARATH; Transcription factor UNE10 | ||||
| TrEMBL | A0A151RAT6 | 0.0 | A0A151RAT6_CAJCA; Transcription factor UNE10 | ||||
| STRING | XP_007140812.1 | 0.0 | (Phaseolus vulgaris) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF5676 | 33 | 47 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G00050.1 | 1e-102 | bHLH family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



