 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | CA01g15440 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Solanoideae; Capsiceae; Capsicum
|
||||||||
| Family | EIL | ||||||||
| Protein Properties | Length: 678aa MW: 76068.6 Da PI: 6.0115 | ||||||||
| Description | EIL family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | EIN3 | 423.1 | 3e-129 | 40 | 388 | 1 | 335 |
XXXXXXXXXXXXXXXXXXXXXXX..XXXXX.XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX CS
EIN3 1 eelkkrmwkdqmllkrlkerkkqlledkeaatgakksnksneqarrkkmsraQDgiLkYMlkemevcnaqGfvYgiipekgkpvegasdsLraWWkekv 99
eel++rmwkd+++lkrlker+k +++ a++++k++++++qarrkkmsraQDgiLkYMlk mevc+a+GfvYgiipekgkpv+gasd++raWWkekv
CA01g15440 40 EELERRMWKDRVKLKRLKERQKLA--AQQ-AAEKQKNKQTTDQARRKKMSRAQDGILKYMLKLMEVCKARGFVYGIIPEKGKPVSGASDNIRAWWKEKV 135
8*******************9963..354.599****************************************************************** PP
XXXXXXXXXXXXXXXXXXXXXXXXXX....XX----STTS-HHHHHHHHHHHSSSSSS-TTS--TTT--HHHH---S--HHHHHHT--TT--.-----G CS
EIN3 100 efdrngpaaiskyqaknlilsgesslqtersseshslselqDTtlgSLLsalmqhcdppqrrfplekgvepPWWPtGkelwwgelglskdqgtppykkp 198
+fd+ngpaai+ky+a++ + +++ ++ ++ + ++ l++lqD+tlgSLLs+lmqhcdppqr++plekg++pPWWP+G+e+ww+++gl+k q+ ppykkp
CA01g15440 136 KFDKNGPAAIAKYEAECYAKGEGIGN--QNGNPQSVLQDLQDATLGSLLSSLMQHCDPPQRKYPLEKGISPPWWPSGNEEWWAKMGLPKGQN-PPYKKP 231
******************99988888..56999999********************************************************.9***** PP
GG--HHHHHHHHHHHHHHTGGGHHHHHHTTTTSSSSTTT--SHHHHHHHHHHTTTTT-S--XXXX..XX.......XXXX..XXXXXXXXXXXXXXXX. CS
EIN3 199 hdlkkawkvsvLtavikhmsptieeirelerqskylqdkmsakesfallsvlnqeekecatvsah..ss.......slrk..qspkvtlsceqkedve. 285
hdlkk++kv+vLtavikhmsp+i++ir+l+rqsk+lqdkm+akes ++l+vl++ee+ +++ s++ ss +++k +++++++dv+
CA01g15440 232 HDLKKMFKVAVLTAVIKHMSPDIAKIRRLVRQSKCLQDKMTAKESSIWLAVLSREESTIQQPSSDngSSsiseaptG--GhgEKKKPSVDSDSDYDVDd 328
*****************************************************************764445555432..14477778888888899996 PP
......XXXXXX.XXXXXXXXXX.................XXXXXXXXXXXXXXXXXXXXX......XXXXXX CS
EIN3 286 ......gkkeskikhvqavktta.................gfpvvrkrkkkpsesakvsskevsrtcqssqfr 335
++ + + +++ +r++++ ++ +++ ++ s+q
CA01g15440 329 vnvsvsS------RDERR----NepsdacpvsavpqshqgKEQGDGQRRRRKCARSNFQQT---QLSPSEQRL 388
5444331......11222....112234444434433332222222222222222222222...333444444 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF04873 | 1.8E-124 | 40 | 287 | No hit | No description |
| Gene3D | G3DSA:1.10.3180.10 | 1.6E-69 | 164 | 295 | IPR023278 | Ethylene insensitive 3-like protein, DNA-binding domain |
| SuperFamily | SSF116768 | 4.18E-60 | 168 | 290 | IPR023278 | Ethylene insensitive 3-like protein, DNA-binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0042762 | Biological Process | regulation of sulfur metabolic process | ||||
| GO:0071281 | Biological Process | cellular response to iron ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 678 aa Download sequence Send to blast |
MEYCMIDADG LGNSSDIEVD DIRCDNIAEK DVSDEEIDPE ELERRMWKDR VKLKRLKERQ 60 KLAAQQAAEK QKNKQTTDQA RRKKMSRAQD GILKYMLKLM EVCKARGFVY GIIPEKGKPV 120 SGASDNIRAW WKEKVKFDKN GPAAIAKYEA ECYAKGEGIG NQNGNPQSVL QDLQDATLGS 180 LLSSLMQHCD PPQRKYPLEK GISPPWWPSG NEEWWAKMGL PKGQNPPYKK PHDLKKMFKV 240 AVLTAVIKHM SPDIAKIRRL VRQSKCLQDK MTAKESSIWL AVLSREESTI QQPSSDNGSS 300 SISEAPTGGH GEKKKPSVDS DSDYDVDDVN VSVSSRDERR NEPSDACPVS AVPQSHQGKE 360 QGDGQRRRRK CARSNFQQTQ LSPSEQRLDD EARNTLPDIN NSNVQFSGYI TNESQPENNM 420 MGPRKPVEKN SGGQSELPLK QSNLSMVPSV NAVSTEDAFV GNRPLIYPML ENSEVVAYES 480 RFHLGTQDSA VQLQLHDTQL QNRPQFSGMN NERRSSIFHY GLPNNGLYNG PQSSILHHEL 540 QDSSFARGFQ YSNLHQPPVY QYYTASAEFG SAHEEQQSHL AFGQPQIKPQ DSGVKSAVLP 600 GDRNDISRED HHYGKDTFQN DHDRPVEMQF ASPITSGSPD YARLSSPFNF DLDVPNTLDP 660 GDLELFLDED VMTFFAS* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wij_A | 7e-72 | 167 | 298 | 9 | 140 | ETHYLENE-INSENSITIVE3-like 3 protein |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that may be involved in the ethylene response pathway. {ECO:0000269|PubMed:9215635, ECO:0000269|PubMed:9851977}. | |||||
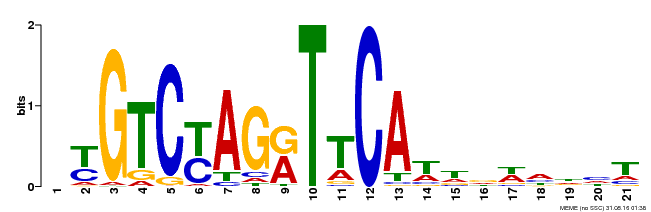
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00233 | DAP | Transfer from AT1G73730 | Download |
| |||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | HG975440 | 0.0 | HG975440.1 Solanum pennellii chromosome ch01, complete genome. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_016556227.1 | 0.0 | PREDICTED: ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| Refseq | XP_016556228.1 | 0.0 | PREDICTED: ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| Swissprot | O23116 | 1e-180 | EIL3_ARATH; ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| TrEMBL | A0A2G3AJ97 | 0.0 | A0A2G3AJ97_CAPAN; ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| STRING | PGSC0003DMT400076890 | 0.0 | (Solanum tuberosum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA304 | 23 | 178 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G73730.1 | 1e-162 | ETHYLENE-INSENSITIVE3-like 3 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



