 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | CA02g05990 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Solanoideae; Capsiceae; Capsicum
|
||||||||
| Family | SRS | ||||||||
| Protein Properties | Length: 363aa MW: 39634.9 Da PI: 7.2327 | ||||||||
| Description | SRS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF702 | 222.6 | 8.1e-69 | 140 | 288 | 3 | 154 |
DUF702 3 sgtasCqdCGnqakkdCaheRCRtCCksrgfdCathvkstWvpaakrrerqqqlaaasskaaasaaeaaskrkrelkskkqsalsstklssaeskkele 101
g++sCqdCGnqakkdC+h+RCRtCCksrgf+C+thvkstWvpaakrrerqqql a++++ + ++++++k+kr+ + ++s+l s++l+s+++ le
CA02g05990 140 GGGISCQDCGNQAKKDCQHMRCRTCCKSRGFQCQTHVKSTWVPAAKRRERQQQLMALQQEGHN-NNNNNNKSKRQREDPSTSSLVSSRLPSTTTG--LE 235
5789*************************************************9998766544.4444445555555588899999999999765..89 PP
DUF702 102 tsslPeevsseavfrcvrvssvddgeeelaYqtavsigGhvfkGiLydqGlee 154
+ ++P++vs++avf+c+++ss+dd+e++laYq+avsigGhvfkGiLydqG e+
CA02g05990 236 VGKFPAKVSTSAVFQCIQMSSIDDAEDQLAYQAAVSIGGHVFKGILYDQGHES 288
999***********************************************987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF05142 | 3.2E-65 | 142 | 287 | IPR007818 | Protein of unknown function DUF702 |
| TIGRFAMs | TIGR01623 | 2.0E-27 | 144 | 186 | IPR006510 | Zinc finger, lateral root primordium type 1 |
| TIGRFAMs | TIGR01624 | 2.1E-24 | 238 | 285 | IPR006511 | Lateral Root Primordium type 1, C-terminal |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010051 | Biological Process | xylem and phloem pattern formation | ||||
| GO:0010252 | Biological Process | auxin homeostasis | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0048479 | Biological Process | style development | ||||
| GO:0048480 | Biological Process | stigma development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0046982 | Molecular Function | protein heterodimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 363 aa Download sequence Send to blast |
MANFFSLGAG SGNQEQQQQQ EISSCHRSNN HNPAVPTEIS QESWFLYRND HNNQEIPTTY 60 KGFELWQSDT TTQHQPEQNQ HQQQHQFRHP IYPLQDFYST AVGLGVGPSR SGFDLSADPT 120 LYAGDHEASR SGFMMMRSGG GGISCQDCGN QAKKDCQHMR CRTCCKSRGF QCQTHVKSTW 180 VPAAKRRERQ QQLMALQQEG HNNNNNNNKS KRQREDPSTS SLVSSRLPST TTGLEVGKFP 240 AKVSTSAVFQ CIQMSSIDDA EDQLAYQAAV SIGGHVFKGI LYDQGHESQY NNMSAAGGDT 300 SSGGSAGVQN HHNSTAVAXX GGAGTAVRPT EASNFHDPSL FPTPLSTFMV AGTQFFPPSR 360 SP* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 208 | 213 | KSKRQR |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator involved in the transcriptional regulation of terpene biosynthesis in glandular trichomes (PubMed:24884371, PubMed:24142382). Binds to the promoter of the linalool synthase TPS5 and promotes TPS5 gene transactivation (PubMed:24884371, PubMed:24142382). Acts synergistically with MYC1 in the transactivation of TPS5 (PubMed:24884371). {ECO:0000269|PubMed:24142382, ECO:0000269|PubMed:24884371}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00613 | PBM | Transfer from AT3G51060 | Download |
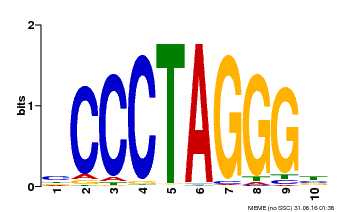
| |||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KC331910 | 0.0 | KC331910.1 Solanum lycopersicum expression of terpenoids 1 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_016561846.1 | 0.0 | PREDICTED: protein SHI RELATED SEQUENCE 1-like isoform X1 | ||||
| Swissprot | K4B6C9 | 1e-169 | EOT1_SOLLC; Protein EXPRESSION OF TERPENOIDS 1 | ||||
| TrEMBL | A0A1U8FRM0 | 0.0 | A0A1U8FRM0_CAPAN; protein SHI RELATED SEQUENCE 1-like isoform X1 | ||||
| STRING | PGSC0003DMT400040107 | 1e-179 | (Solanum tuberosum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA678 | 24 | 110 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G66350.1 | 3e-67 | Lateral root primordium (LRP) protein-related | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



