 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | CA02g07170 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Solanoideae; Capsiceae; Capsicum
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 494aa MW: 53990.3 Da PI: 5.897 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 48.1 | 2.8e-15 | 49 | 93 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
r +WT+eE++++++a k++G W+ I +++g +t+ q++s+ qk+
CA02g07170 49 REKWTEEEHQRFLEALKLYGRA-WRQIEEYVG-SKTAIQIRSHAQKF 93
789*****************88.*********.************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 9.72E-16 | 43 | 97 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 21.179 | 44 | 98 | IPR017930 | Myb domain |
| TIGRFAMs | TIGR01557 | 2.6E-17 | 47 | 96 | IPR006447 | Myb domain, plants |
| SMART | SM00717 | 3.6E-12 | 48 | 96 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.6E-8 | 49 | 89 | IPR009057 | Homeodomain-like |
| Pfam | PF00249 | 1.9E-12 | 49 | 92 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 5.30E-9 | 51 | 94 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0007623 | Biological Process | circadian rhythm | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 494 aa Download sequence Send to blast |
NQAEGAWQGA SSVNSSKLEI VATQEKAPTC LANKNALKVR KPYTITKQRE KWTEEEHQRF 60 LEALKLYGRA WRQIEEYVGS KTAIQIRSHA QKFFAKVARD SVNDGDESLN AIDIPPPRPK 120 KKPLHPYPRK MVDSPVANKA VSGQPESSPS PNVSGRVSRS PDSVLSAIGS GVSEYPVAEQ 180 QNSRFSPASC TTDVHTANII SAENDDESMT SNSSTVEEVH VELKPISAST SPIANSDMVI 240 PLEACNLSAL PCYLGCTLTS GLYVPLQECD IVHRENSCNG EKLAVEAPSA SIKLFGQTVF 300 IPDANNLALR APYNCNSLPS KSTEKEIKIS SEDVLHSFQA NQANSQFMLA MVPGNMIPPA 360 SWLSHNMLEN NPEITTVFPT TVSWWSWYQD LVNRSILSCD QKAMETAVHC QGPKDEEPQR 420 EGSSTGSSIG SASEVDDGNR SSETAESKCT AKSYKWNNSK GFVPYKRCLA ERDDKSSGAV 480 LEERESQRVR VCS* |
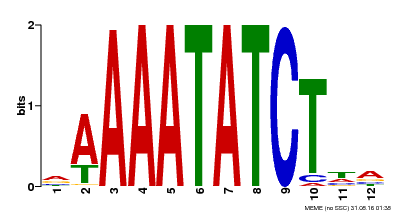
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00146 | DAP | Transfer from AT1G18330 | Download |
| |||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AC151804 | 0.0 | AC151804.1 Solanum demissum chromosome 5 clone PGEC475B22, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_016563966.1 | 0.0 | PREDICTED: protein REVEILLE 1-like isoform X1 | ||||
| TrEMBL | A0A1U8FXT5 | 0.0 | A0A1U8FXT5_CAPAN; protein REVEILLE 1-like isoform X1 | ||||
| STRING | PGSC0003DMT400065850 | 0.0 | (Solanum tuberosum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA7447 | 21 | 27 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G18330.1 | 5e-40 | MYB_related family protein | ||||



