 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | CA02g12980 | ||||||||
| Common Name | AGL2 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Solanoideae; Capsiceae; Capsicum
|
||||||||
| Family | MIKC_MADS | ||||||||
| Protein Properties | Length: 248aa MW: 28634.3 Da PI: 9.8599 | ||||||||
| Description | MIKC_MADS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SRF-TF | 99.3 | 1.6e-31 | 25 | 74 | 1 | 50 |
S---SHHHHHHHHHHHHHHHHHHHHHHHHHHT-EEEEEEE-TTSEEEEEE CS
SRF-TF 1 krienksnrqvtfskRrngilKKAeELSvLCdaevaviifsstgklyeys 50
krien++nrqvtf+kRrng+lKKA+ELSvLCdaeva+i+fss+g+lyey+
CA02g12980 25 KRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYA 74
79***********************************************8 PP
| |||||||
| 2 | K-box | 112.6 | 4e-37 | 93 | 189 | 4 | 100 |
K-box 4 ssgksleeakaeslqqelakLkkeienLqreqRhllGedLesLslkeLqqLeqqLekslkkiRskKnellleqieelqkkekelqeenkaLrkklee 100
s++ s +ea+a+++qqe++kL+++i nLq+++R+++Ge L L+++eL++Leq++ek+++k+RskKne+l+++ie++qk+e +l+++n++Lr+k++e
CA02g12980 93 SNTGSISEANAQYYQQESSKLRAQIGNLQNQNRNYMGEALAALNHRELRNLEQKIEKGISKVRSKKNEMLFAEIEYMQKREVDLHNNNQYLRAKIAE 189
4444599***************************************************************************************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00432 | 3.4E-41 | 17 | 76 | IPR002100 | Transcription factor, MADS-box |
| PROSITE profile | PS50066 | 33.669 | 17 | 77 | IPR002100 | Transcription factor, MADS-box |
| CDD | cd00265 | 4.14E-46 | 18 | 94 | No hit | No description |
| SuperFamily | SSF55455 | 3.27E-33 | 18 | 90 | IPR002100 | Transcription factor, MADS-box |
| PROSITE pattern | PS00350 | 0 | 19 | 73 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 4.6E-33 | 19 | 39 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF00319 | 9.3E-27 | 26 | 73 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 4.6E-33 | 39 | 54 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 4.6E-33 | 54 | 75 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF01486 | 4.9E-26 | 101 | 187 | IPR002487 | Transcription factor, K-box |
| PROSITE profile | PS51297 | 14.836 | 103 | 193 | IPR002487 | Transcription factor, K-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0048366 | Biological Process | leaf development | ||||
| GO:0048440 | Biological Process | carpel development | ||||
| GO:0048443 | Biological Process | stamen development | ||||
| GO:0048497 | Biological Process | maintenance of floral organ identity | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 248 aa Download sequence Send to blast |
MDFQSDLTRE ISPQRKLGRG KIEIKRIENT TNRQVTFCKR RNGLLKKAYE LSVLCDAEVA 60 LIVFSSRGRL YEYANNSVKA TIERYKKACS DSSNTGSISE ANAQYYQQES SKLRAQIGNL 120 QNQNRNYMGE ALAALNHREL RNLEQKIEKG ISKVRSKKNE MLFAEIEYMQ KREVDLHNNN 180 QYLRAKIAET ERAQQHQQQM NLMPGSSSSY ELVPPPQQFD TRNYLQVNGL QTNNHYPRQD 240 QPPLQLV* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1tqe_P | 1e-20 | 17 | 85 | 1 | 69 | Myocyte-specific enhancer factor 2B |
| 1tqe_Q | 1e-20 | 17 | 85 | 1 | 69 | Myocyte-specific enhancer factor 2B |
| 1tqe_R | 1e-20 | 17 | 85 | 1 | 69 | Myocyte-specific enhancer factor 2B |
| 1tqe_S | 1e-20 | 17 | 85 | 1 | 69 | Myocyte-specific enhancer factor 2B |
| 5f28_A | 1e-20 | 17 | 85 | 1 | 69 | MEF2C |
| 5f28_B | 1e-20 | 17 | 85 | 1 | 69 | MEF2C |
| 5f28_C | 1e-20 | 17 | 85 | 1 | 69 | MEF2C |
| 5f28_D | 1e-20 | 17 | 85 | 1 | 69 | MEF2C |
| 6byy_A | 1e-20 | 17 | 85 | 1 | 69 | MEF2 CHIMERA |
| 6byy_B | 1e-20 | 17 | 85 | 1 | 69 | MEF2 CHIMERA |
| 6byy_C | 1e-20 | 17 | 85 | 1 | 69 | MEF2 CHIMERA |
| 6byy_D | 1e-20 | 17 | 85 | 1 | 69 | MEF2 CHIMERA |
| 6bz1_A | 2e-20 | 17 | 85 | 1 | 69 | MEF2 CHIMERA |
| 6bz1_B | 2e-20 | 17 | 85 | 1 | 69 | MEF2 CHIMERA |
| 6c9l_A | 1e-20 | 17 | 85 | 1 | 69 | Myocyte-specific enhancer factor 2B |
| 6c9l_B | 1e-20 | 17 | 85 | 1 | 69 | Myocyte-specific enhancer factor 2B |
| 6c9l_C | 1e-20 | 17 | 85 | 1 | 69 | Myocyte-specific enhancer factor 2B |
| 6c9l_D | 1e-20 | 17 | 85 | 1 | 69 | Myocyte-specific enhancer factor 2B |
| 6c9l_E | 1e-20 | 17 | 85 | 1 | 69 | Myocyte-specific enhancer factor 2B |
| 6c9l_F | 1e-20 | 17 | 85 | 1 | 69 | Myocyte-specific enhancer factor 2B |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor involved in regulating genes that determines stamen and carpel development in wild-type flowers. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00609 | ChIP-seq | Transfer from AT4G18960 | Download |
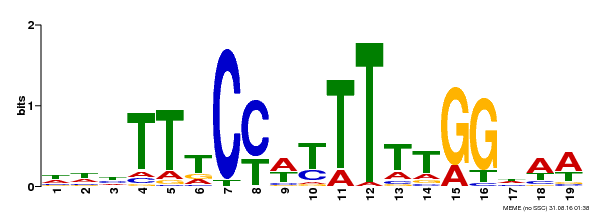
| |||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | HM565943 | 0.0 | HM565943.1 Capsicum annuum AGAMOUS-LIKE2 (AGL2) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | NP_001311689.1 | 0.0 | floral homeotic protein AGAMOUS | ||||
| Swissprot | Q40168 | 1e-170 | AG_SOLLC; Floral homeotic protein AGAMOUS | ||||
| TrEMBL | A0A1U8FVA7 | 0.0 | A0A1U8FVA7_CAPAN; floral homeotic protein AGAMOUS isoform X2 | ||||
| TrEMBL | A0A2G3D4W2 | 0.0 | A0A2G3D4W2_CAPCH; Floral homeotic protein AGAMOUS | ||||
| TrEMBL | E3UT60 | 0.0 | E3UT60_CAPAN; AGAMOUS-LIKE2 | ||||
| STRING | PGSC0003DMT400073134 | 1e-170 | (Solanum tuberosum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA40 | 24 | 625 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G18960.1 | 1e-123 | MIKC_MADS family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



