 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | CA02g18810 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Solanoideae; Capsiceae; Capsicum
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 490aa MW: 53548.9 Da PI: 8.3152 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 49 | 1.4e-15 | 56 | 100 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
r rWT+eE++++++a k++G W++I +++ ++t+ q++s+ qk+
CA02g18810 56 RERWTEEEHKKFLEALKLYGRA-WRRIEEHVS-TKTAVQIRSHAQKF 100
78******************88.*********.************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 20.28 | 51 | 105 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 4.11E-16 | 51 | 104 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 1.5E-16 | 54 | 103 | IPR006447 | Myb domain, plants |
| SMART | SM00717 | 2.2E-12 | 55 | 103 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 7.8E-13 | 56 | 99 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.0E-9 | 56 | 96 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 9.35E-10 | 58 | 101 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0007623 | Biological Process | circadian rhythm | ||||
| GO:0009734 | Biological Process | auxin-activated signaling pathway | ||||
| GO:0010600 | Biological Process | regulation of auxin biosynthetic process | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 490 aa Download sequence Send to blast |
DQSGESGVDS IHSSRNQISS DVGASSTRSI KLKEQVASAD EYAPKVRKPY IITKQRERWT 60 EEEHKKFLEA LKLYGRAWRR IEEHVSTKTA VQIRSHAQKF FSKVVRESNN GDANSVKPIE 120 IPPPRPKRKP MHPYPRKLAT PVIIGTPVPE KSTRSLTPNV SMSEQENQSP TSVLSVHGSD 180 ALGTVDSSKR LGSASPISSA GDGNGNSGDF VLSEPPNSIL EDSSSPVQAN AKTNPENQTF 240 VKLELFPQDD DFVQEGSAET SSTQCLKLFG KTVLFTESQK PSCPTPGPCK IETGLNDEAS 300 LPAVLWNVMP MKFTAADSEC ASRTLTLGTP SPFYILPSQN ENKRPTGCAS STLLPWGSSC 360 ASASFSCIQV LNPIPVKGRP IFNDRDLEDK QNQKEGSSTG SNTEPVSAET GGEKNMDIET 420 QSSRHPLEKM GKDRSSFRPR ETFSLVQRAS STKRIKGFIP YKRCLTESGI NSTTLSGEER 480 EEQRTRLCL* |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00515 | DAP | Transfer from AT5G17300 | Download |
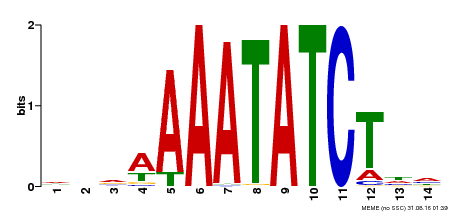
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_016461216.1 | 0.0 | PREDICTED: protein REVEILLE 1-like isoform X1 | ||||
| TrEMBL | A0A2G3A9B2 | 0.0 | A0A2G3A9B2_CAPAN; Protein REVEILLE 2 | ||||
| STRING | XP_009613640.1 | 0.0 | (Nicotiana tomentosiformis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA3185 | 24 | 47 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G17300.1 | 2e-60 | MYB_related family protein | ||||



