 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | CA03g25230 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Solanoideae; Capsiceae; Capsicum
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 1273aa MW: 141303 Da PI: 9.065 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 11 | 0.0014 | 1180 | 1203 | 2 | 23 |
EET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 2 kCp..dCgksFsrksnLkrHirtH 23
+Cp C+k F ++ +L++H r+H
CA03g25230 1180 VCPveGCKKKFFSHKYLVQHRRVH 1203
69999*****************99 PP
| |||||||
| 2 | zf-C2H2 | 11.9 | 0.00071 | 1239 | 1265 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH..T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt..H 23
y+C Cg++F+ s++ rH r+ H
CA03g25230 1239 YVCAetGCGQTFRFVSDFSRHKRKtgH 1265
89********************99666 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00545 | 1.0E-15 | 19 | 60 | IPR003349 | JmjN domain |
| PROSITE profile | PS51183 | 14.395 | 20 | 61 | IPR003349 | JmjN domain |
| Pfam | PF02375 | 2.8E-15 | 21 | 54 | IPR003349 | JmjN domain |
| SMART | SM00558 | 9.0E-49 | 187 | 356 | IPR003347 | JmjC domain |
| PROSITE profile | PS51184 | 32.57 | 190 | 356 | IPR003347 | JmjC domain |
| SuperFamily | SSF51197 | 2.61E-25 | 202 | 371 | No hit | No description |
| Pfam | PF02373 | 1.1E-36 | 220 | 339 | IPR003347 | JmjC domain |
| SMART | SM00355 | 7.4 | 1156 | 1178 | IPR015880 | Zinc finger, C2H2-like |
| SMART | SM00355 | 0.012 | 1179 | 1203 | IPR015880 | Zinc finger, C2H2-like |
| Gene3D | G3DSA:3.30.160.60 | 4.4E-5 | 1179 | 1207 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 11.572 | 1179 | 1208 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 1181 | 1203 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 1.4E-9 | 1195 | 1237 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 2.9E-8 | 1208 | 1233 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 10.741 | 1209 | 1238 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.0014 | 1209 | 1233 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 1211 | 1233 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 8.1E-8 | 1227 | 1261 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 1.3E-8 | 1234 | 1261 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 0.85 | 1239 | 1265 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 10.907 | 1239 | 1270 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 1241 | 1265 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009741 | Biological Process | response to brassinosteroid | ||||
| GO:0009826 | Biological Process | unidimensional cell growth | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0033169 | Biological Process | histone H3-K9 demethylation | ||||
| GO:0035067 | Biological Process | negative regulation of histone acetylation | ||||
| GO:0048366 | Biological Process | leaf development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003676 | Molecular Function | nucleic acid binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1273 aa Download sequence Send to blast |
MAEASGNIEV FPWLKTLPVA PEYYPTLEEF QDPIAYIFKI EKEASKYGIC KIVPPLPAPK 60 QKTALANLNR SLSARPGSNG PTFTTRQQQI GFCPRKHRPV KKPVWQSGET YTVQQFQAKA 120 KAFEKNYLKR NSKRALTALE IETLYWKATV DKPFSVEYAN DMPGSAFAPK KAGSGSVGAG 180 GIGEVATLAD TEWNMRGVSR SKGSLLKFMK EEIPGVTSPM VYLAMMFSWF AWHVEDHDLH 240 SLNYLHMGAG KTWYGVPRDA AVAFEEVISV QGYAGETNPL VTFATLGEKT TVMSPEVLLS 300 AGIPCCRLVQ NAGDFVVTFP RAYHSGFSHG FNCGEASNIA TPEWLRFAKD AAIRRASTNC 360 PPMVSHFQLL YDLALSLCSR VPKNIRIEPR SSRLKDKKKS EGDMLVKELF VEDLNSNNYL 420 LHVLGEGSPV VLLPQTSTGI SICSNLVAGT QSKVNSRLFL GLSSSDHELR SQKDIGYDHF 480 KLGRKQGMKQ VASVSSEKGK YSSLHAGNRL PDSGRKDDAQ SSPETERVNL DTGRGMTHKC 540 DTLSEQGLFS CVTCGILCYT CVAIIRPTEA AAHHLMSSDY HNFNDWTGNV GGVFATGRDP 600 NAGESDSSSG GLVKTAPGDF IDAPVESSDR IQKLNNGSVE GVSRTKARKE TSSLGLLALA 660 YANSSDSDED EVEADIPVEA CESRHTDSDD EVFLQVIDPY ANHRQRRAVS QGRSCQKIDN 720 SSPSGESNTL FGRSSHQLRS NQVTAKCISN KGEIAQNNAV APFDNTHMQF TSASDEDSFR 780 IHVFCLQHAV QVEEKLRQIG GADISLLCHP DYPKLEAQAK KVAEELGSDH FWRDISFREA 840 TKEDEEMIQS ALEIQEAIHG NGDWTVKLDI NLFYSANLSR SPLYSKQMPY NFIIYNAFGR 900 DSPDNTPEKS EYTGRGSGKQ RRAVVAGKWC GKVWMSSQVH PLLAERDIDE EQEQNKNISV 960 QIKTEVKSER PRERTVTGKT VATTCKTGKK RNSTAEDRNA SNTQLIADDH DDSLLCSLLQ 1020 QHRKTNFRSK RIKYETPEPQ KDVDKKKRFG SLIDDDPDGG PSARLNVDKK KRFGSLIDDD 1080 PDGGPSTRLR KRVPKPSKVS AAKLIKAKPA TTKQEESKKG PKVKLPSVNS TAKKEPVMKS 1140 PRSNTGRKMR DKEGEYHCDL EGCSMSFSSK PELTLHKKNV CPVEGCKKKF FSHKYLVQHR 1200 RVHMDDRPLK CPWKGCKMTF KWAWARTEHI RVHTGARPYV CAETGCGQTF RFVSDFSRHK 1260 RKTGHVSKKG RG* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6ip0_A | 4e-73 | 13 | 389 | 8 | 366 | Transcription factor jumonji (Jmj) family protein |
| 6ip4_A | 4e-73 | 13 | 389 | 8 | 366 | Arabidopsis JMJ13 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Histone demethylase that demethylates 'Lys-27' (H3K27me) of histone H3. Demethylates both tri- (H3K27me3) and di-methylated (H3K27me2) H3K27me (PubMed:21642989, PubMed:27111035). Demethylates also H3K4me3/2 and H3K36me3/2 in an in vitro assay (PubMed:20711170). Involved in the transcriptional regulation of hundreds of genes regulating developmental patterning and responses to various stimuli (PubMed:18467490). Binds DNA via its four zinc fingers in a sequence-specific manner, 5'-CTCTGYTY-3', to promote the demethylation of H3K27me3 and the regulation of organ boundary formation (PubMed:27111034, PubMed:27111035). Involved in the regulation of flowering time by repressing FLOWERING LOCUS C (FLC) expression (PubMed:15377760). Interacts with the NF-Y complexe to regulate SOC1 (PubMed:25105952). Mediates the recruitment of BRM to its target loci (PubMed:27111034). {ECO:0000269|PubMed:15377760, ECO:0000269|PubMed:18467490, ECO:0000269|PubMed:20711170, ECO:0000269|PubMed:21642989, ECO:0000269|PubMed:25105952, ECO:0000269|PubMed:27111034, ECO:0000269|PubMed:27111035}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00608 | ChIP-seq | Transfer from AT3G48430 | Download |
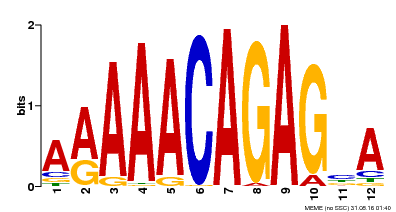
| |||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | HG975442 | 0.0 | HG975442.1 Solanum pennellii chromosome ch03, complete genome. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_016563761.1 | 0.0 | PREDICTED: lysine-specific demethylase REF6 | ||||
| Swissprot | Q9STM3 | 0.0 | REF6_ARATH; Lysine-specific demethylase REF6 | ||||
| TrEMBL | A0A1U8FXA8 | 0.0 | A0A1U8FXA8_CAPAN; Lysine-specific demethylase REF6 | ||||
| TrEMBL | A0A2G2ZW03 | 0.0 | A0A2G2ZW03_CAPAN; lysine-specific demethylase REF6 | ||||
| STRING | PGSC0003DMT400039224 | 0.0 | (Solanum tuberosum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA4988 | 23 | 31 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G48430.1 | 0.0 | relative of early flowering 6 | ||||



