 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | CA03g28940 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Solanoideae; Capsiceae; Capsicum
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 542aa MW: 58913.1 Da PI: 7.9721 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 37.1 | 5.6e-12 | 245 | 293 | 2 | 55 |
HHHHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 2 rrahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
r +h++ E+rRR++iN++ + Lr ++P++ ++K +Ka+ L +++eYI+ Lq
CA03g28940 245 RSKHSATEQRRRSKINDR-HMLRGIIPNS----DQKRDKASFLLEVIEYIHFLQ 293
889***************.89*******9....9******************99 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.280.10 | 5.4E-19 | 240 | 309 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 2.22E-14 | 241 | 310 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 10.976 | 243 | 292 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 1.1E-9 | 245 | 293 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 4.36E-11 | 245 | 297 | No hit | No description |
| SMART | SM00353 | 1.8E-8 | 249 | 298 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006351 | Biological Process | transcription, DNA-templated | ||||
| GO:0009742 | Biological Process | brassinosteroid mediated signaling pathway | ||||
| GO:1902448 | Biological Process | positive regulation of shade avoidance | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 542 aa Download sequence Send to blast |
MYLISPFSMS VVHANKEGGY LKTHDFLQPL EQAEMTSRKE DTKVEVVAVE RPSPPLAATP 60 SGEHTLPGGI GTFSISYLHQ RVPKPEASLF SVAQASSTDR NDENSNCSSY TGSGFTLWDE 120 SAVNKGKTGK ENSGGDRHIL REAGVNTGGV HPTTSLEWQS QSSSNHKHNT AALSSLSSAQ 180 QSSPLKSQNF LHMITSAKNA QDDDDDEDED CVIKKEPQSH PRGLSCNLSV KVDGKGNDQK 240 PNTPRSKHSA TEQRRRSKIN DRHMLRGIIP NSDQKRDKAS FLLEVIEYIH FLQGKVHKYE 300 ESYQGWDNEP LKLPLSKCHR TTHGVSNHPQ CTINASGAAV PYARKFDESV MAISSANPIN 360 VQKLEPNIST TSLKENGQQP GLTNKATALP MHPNTFSFCG TNSTAALHSS KLMADTDKLE 420 SKSHSEFSLS RSHMTDYAIP NTNPKGQALS IESGTISISS AYSQGLLNTL TQALQNSGVD 480 LSQANISVQI DLGKRANGRV NSSASTIKGD DVSTSNQPTP QSRVTTTREE SDQAFKRRKT 540 S* |
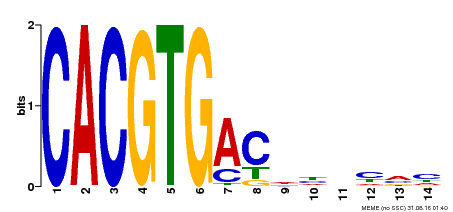
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00498 | DAP | Transfer from AT5G08130 | Download |
| |||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT013786 | 0.0 | BT013786.1 Lycopersicon esculentum clone 132679F, mRNA sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_016563392.1 | 0.0 | PREDICTED: transcription factor BIM1 isoform X1 | ||||
| TrEMBL | A0A2G3A1U9 | 0.0 | A0A2G3A1U9_CAPAN; Transcription factor BIM3 | ||||
| STRING | Solyc03g114720.2.1 | 0.0 | (Solanum lycopersicum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA3969 | 23 | 45 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G08130.4 | 4e-83 | bHLH family protein | ||||



