 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | CA04g11250 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Solanoideae; Capsiceae; Capsicum
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 517aa MW: 57630.3 Da PI: 6.6038 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 21 | 8.6e-07 | 264 | 285 | 2 | 23 |
EETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 2 kCpdCgksFsrksnLkrHirtH 23
C++Cgk F+r nL+ H+r H
CA04g11250 264 FCTICGKGFKRDANLRMHMRGH 285
6*******************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF57667 | 2.32E-5 | 261 | 288 | No hit | No description |
| PROSITE profile | PS50157 | 12.03 | 263 | 290 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.0026 | 263 | 285 | IPR015880 | Zinc finger, C2H2-like |
| Gene3D | G3DSA:3.30.160.60 | 8.3E-6 | 264 | 314 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE pattern | PS00028 | 0 | 265 | 285 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 62 | 312 | 345 | IPR015880 | Zinc finger, C2H2-like |
| SMART | SM00355 | 25 | 350 | 372 | IPR015880 | Zinc finger, C2H2-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010044 | Biological Process | response to aluminum ion | ||||
| GO:0010447 | Biological Process | response to acidic pH | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003676 | Molecular Function | nucleic acid binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 517 aa Download sequence Send to blast |
MDPDDSLTED PWIKSSSSGN ELLKIMPSDN HSFTNFNLHA QKWEGSSYLD QQIRIEQQFS 60 GFTKPKHTSE MDQQGNHRNE NHDSTRIRDW DPRTLLNNLS FLEQKIHQLQ ELVHLIVGRR 120 GQDGVQGNDL IVQQQQLITA DLTSIIVQLI STAGSLLPTM KHTLSSASPA ANQLGQVGGV 180 TIPSTIGTSA SAGGLTCNDG VAKVEDQSNH VDQLRDCGIE QNHPADEHEL KDEDEAEEEE 240 NLPPGSYEIL QLEKEEILAP HTHFCTICGK GFKRDANLRM HMRGHGDEYK TPAALAKPHK 300 EPSSEPTLIK RYSCPYVGCK RNKEHKKFQP LKTILCVKNH YKRTHCEKAY TCSRCNIKKF 360 SVIADLKTHE KHCGKDKWLC SCGTTFSRKD KLFGHISLFQ GHTPAVSPDE TKGSAGMSDR 420 GQTSEVTMKA RQENFKVNAS HGNEFQNPGV VKEGAYGSPS SYFSPLNFDT SNLNGFQEFP 480 RPPFDESDSS LSFLLSGSCE YPPHKAAKYM SSTEME* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. Together with STOP2, plays a critical role in tolerance to major stress factors in acid soils such as proton H(+) and aluminum ion Al(3+). Required for the expression of genes in response to acidic stress (e.g. ALMT1 and MATE), and Al-activated citrate exudation. {ECO:0000269|PubMed:17535918, ECO:0000269|PubMed:18826429, ECO:0000269|PubMed:19321711, ECO:0000269|PubMed:23935008}. | |||||
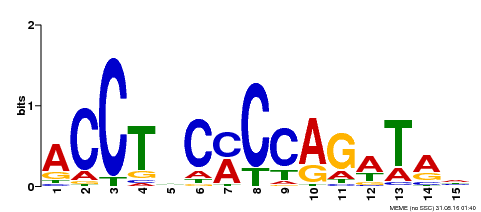
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00196 | ampDAP | Transfer from AT1G34370 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By shock H(+) and Al(3+) treatments. {ECO:0000269|PubMed:17535918}. | |||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | HG975523 | 0.0 | HG975523.1 Solanum lycopersicum chromosome ch11, complete genome. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_016569747.1 | 0.0 | PREDICTED: protein SENSITIVE TO PROTON RHIZOTOXICITY 1 isoform X1 | ||||
| Refseq | XP_016569748.1 | 0.0 | PREDICTED: protein SENSITIVE TO PROTON RHIZOTOXICITY 1 isoform X1 | ||||
| Swissprot | Q9C8N5 | 0.0 | STOP1_ARATH; Protein SENSITIVE TO PROTON RHIZOTOXICITY 1 | ||||
| TrEMBL | A0A1U8GEJ9 | 0.0 | A0A1U8GEJ9_CAPAN; protein SENSITIVE TO PROTON RHIZOTOXICITY 1 isoform X1 | ||||
| STRING | PGSC0003DMT400023881 | 0.0 | (Solanum tuberosum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA4754 | 24 | 41 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G34370.2 | 1e-171 | C2H2 family protein | ||||



