 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | CA04g14920 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Solanoideae; Capsiceae; Capsicum
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 319aa MW: 35340.5 Da PI: 9.0106 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 63.7 | 3.8e-20 | 182 | 231 | 2 | 55 |
AP2 2 gykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
y+GVr+++ +g+WvAeIr p++ r+r +lg+f+taeeAa a+++a+ l+g
CA04g14920 182 LYRGVRQRH-WGKWVAEIRLPKN---RTRLWLGTFDTAEEAALAYDKAAYMLRG 231
59******9.**********965...5**********************99987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00018 | 7.82E-32 | 181 | 238 | No hit | No description |
| SuperFamily | SSF54171 | 1.11E-22 | 182 | 240 | IPR016177 | DNA-binding domain |
| Gene3D | G3DSA:3.30.730.10 | 2.7E-31 | 182 | 240 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 22.3 | 182 | 239 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 9.6E-35 | 182 | 245 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 2.8E-11 | 183 | 194 | IPR001471 | AP2/ERF domain |
| Pfam | PF00847 | 5.5E-14 | 183 | 231 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 2.8E-11 | 205 | 221 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0009611 | Biological Process | response to wounding | ||||
| GO:0009736 | Biological Process | cytokinin-activated signaling pathway | ||||
| GO:0009873 | Biological Process | ethylene-activated signaling pathway | ||||
| GO:0010017 | Biological Process | red or far-red light signaling pathway | ||||
| GO:0045595 | Biological Process | regulation of cell differentiation | ||||
| GO:0071472 | Biological Process | cellular response to salt stress | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 319 aa Download sequence Send to blast |
MAAAMDFWNS TVPLDLQLSS DPVTSGGELM EALEPFMKSV SPPSTSPVNF PPIFSSPSSD 60 LQSFPSFPPP PPNPISYPYT SSFYPTVQPS DDCSTSTEMN SQIFSTGFSS YGMDQQSSSI 120 GLNQLTPTQI EQIQAQINFQ IQQQQQQQMM SYHHHASSTM NFLAPKPVHM KQSGSPPKPT 180 KLYRGVRQRH WGKWVAEIRL PKNRTRLWLG TFDTAEEAAL AYDKAAYMLR GDFARLNFSH 240 LRHNFGEYNP LHSSVDAKLK DICQSLAQGK SIDSKKKKTK GSASASAXXX XXXXXXXXXX 300 XXXXXXXXGL GSYSCRGS* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2gcc_A | 2e-21 | 181 | 247 | 4 | 70 | ATERF1 |
| 3gcc_A | 2e-21 | 181 | 247 | 4 | 70 | ATERF1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probably acts as a transcriptional activator. Binds to the GCC-box pathogenesis-related promoter element. May be involved in the regulation of gene expression by stress factors and by components of stress signal transduction pathways (By similarity). {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00026 | PBM | Transfer from AT1G78080 | Download |
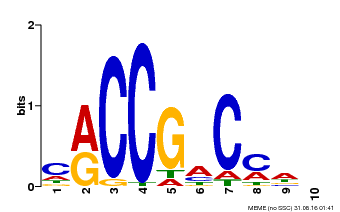
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_016569316.1 | 0.0 | PREDICTED: ethylene-responsive transcription factor RAP2-13-like | ||||
| Swissprot | Q8H1E4 | 1e-66 | RAP24_ARATH; Ethylene-responsive transcription factor RAP2-4 | ||||
| TrEMBL | A0A2G2ZQC4 | 0.0 | A0A2G2ZQC4_CAPAN; Ethylene-responsive transcription factor | ||||
| STRING | PGSC0003DMT400024508 | 1e-121 | (Solanum tuberosum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA1610 | 24 | 71 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G22190.1 | 9e-48 | ERF family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



