 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | CA06g13580 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Solanoideae; Capsiceae; Capsicum
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 580aa MW: 64374.9 Da PI: 7.5576 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 106.2 | 1.6e-33 | 240 | 297 | 2 | 60 |
--SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS-- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhek 60
dDgynWrKYGqK+vkgse+prsYY+Ct+++Cp+kkkvers d++++ei+Y+g+Hnh+k
CA06g13580 240 DDGYNWRKYGQKQVKGSENPRSYYKCTYPNCPTKKKVERSL-DGQITEIVYKGNHNHPK 297
8****************************************.***************85 PP
| |||||||
| 2 | WRKY | 106.6 | 1.3e-33 | 412 | 470 | 1 | 59 |
---SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 1 ldDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhe 59
ldDgy+WrKYGqK+vkg+++prsYY+Cts+gCpv+k+ver+++d k v++tYeg+Hnh+
CA06g13580 412 LDDGYRWRKYGQKVVKGNPNPRSYYKCTSPGCPVRKHVERASQDIKSVITTYEGKHNHD 470
59********************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 4.8E-29 | 231 | 298 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 24.088 | 234 | 298 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 1.09E-25 | 235 | 298 | IPR003657 | WRKY domain |
| SMART | SM00774 | 9.2E-37 | 239 | 297 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 1.3E-25 | 240 | 296 | IPR003657 | WRKY domain |
| Gene3D | G3DSA:2.20.25.80 | 1.5E-37 | 397 | 472 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 5.36E-29 | 404 | 472 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 38.618 | 407 | 472 | IPR003657 | WRKY domain |
| SMART | SM00774 | 1.1E-38 | 412 | 471 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 1.0E-25 | 413 | 470 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0009788 | Biological Process | negative regulation of abscisic acid-activated signaling pathway | ||||
| GO:0009938 | Biological Process | negative regulation of gibberellic acid mediated signaling pathway | ||||
| GO:0010120 | Biological Process | camalexin biosynthetic process | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0010508 | Biological Process | positive regulation of autophagy | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0050832 | Biological Process | defense response to fungus | ||||
| GO:0070370 | Biological Process | cellular heat acclimation | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 580 aa Download sequence Send to blast |
MAASSFSFPT SSSFMTTSFT DLLASSDDYP ITKGLGDRIA ERTGSGVPKF KSLPPPSLPL 60 SPPPFSPSSY FAIPPGLSPT ELLDSPVLLS SSNVLPSPTT GSFPAQAFNW KSSSNNQDVK 120 QEEKNCSDFS FQTQVGTAAS ISQSQTSHVS LVLLPPSQIS CRFILLYMPE ILIKRVSLVT 180 LILCQAWNYQ EPTKQDGLSS DQNANGRSEF NTMQSFMQNN DHSNSGNGYN QSIREQKRSD 240 DGYNWRKYGQ KQVKGSENPR SYYKCTYPNC PTKKKVERSL DGQITEIVYK GNHNHPKPQA 300 TRRSSSSTAS SAIQSYNTQT NEIPDHQSYG SNGTGQIDSV ATPENSSISF GDDDHEHTSQ 360 KSRSRGDDLD EEEPDSKRWK RESESEGLSA LGSRTVREPR VVVQTTSDID ILDDGYRWRK 420 YGQKVVKGNP NPRSYYKCTS PGCPVRKHVE RASQDIKSVI TTYEGKHNHD VPAARGSGNH 480 SINRPIAPTI TNNNSAMAIR PSVTSHQSNY QVPMQSIRPQ QFEMRAPFTL EMLQKPNNYG 540 FSGYANSEDS YENQLQDNNG FSRAKNEPRD DMFMESLLC* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wj2_A | 7e-38 | 223 | 473 | 1 | 78 | Probable WRKY transcription factor 4 |
| 2lex_A | 7e-38 | 223 | 473 | 1 | 78 | Probable WRKY transcription factor 4 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00299 | DAP | Transfer from AT2G38470 | Download |
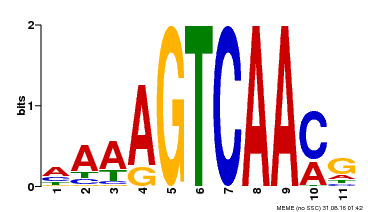
| |||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | DQ402421 | 0.0 | DQ402421.1 Capsicum annuum WRKY-type transcription factor (WRKY2) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | NP_001311944.1 | 0.0 | probable WRKY transcription factor 25 | ||||
| TrEMBL | A0A1U8H1Q5 | 0.0 | A0A1U8H1Q5_CAPAN; Uncharacterized protein | ||||
| TrEMBL | A0A2G2ZAP8 | 0.0 | A0A2G2ZAP8_CAPAN; probable WRKY transcription factor 25 | ||||
| STRING | PGSC0003DMT400043204 | 0.0 | (Solanum tuberosum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA2286 | 24 | 60 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G38470.1 | 1e-112 | WRKY DNA-binding protein 33 | ||||



