 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | CA06g25900 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Solanoideae; Capsiceae; Capsicum
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 465aa MW: 51932.1 Da PI: 8.4509 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 27.1 | 9.5e-09 | 415 | 462 | 3 | 47 |
SS-HHHHHHHHHHHHHTTTT-HHHHHHHHT...TTS-HHHHHHHHHHH CS
Myb_DNA-binding 3 rWTteEdellvdavkqlGggtWktIartmg...kgRtlkqcksrwqky 47
+W++ E++ l +v+++G g+Wk I ++R ++k++w++
CA06g25900 415 KWSALEEDTLRTGVQKYGIGNWKVILDAYRdifTTRQSGDLKDKWRNM 462
8*********************************************96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 10.209 | 408 | 464 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.33E-13 | 410 | 463 | IPR009057 | Homeodomain-like |
| CDD | cd11660 | 8.21E-16 | 414 | 463 | No hit | No description |
| Pfam | PF00249 | 8.6E-7 | 415 | 462 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 3.9E-10 | 415 | 463 | IPR009057 | Homeodomain-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009901 | Biological Process | anther dehiscence | ||||
| GO:0010152 | Biological Process | pollen maturation | ||||
| GO:0043067 | Biological Process | regulation of programmed cell death | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 465 aa Download sequence Send to blast |
MGIDTHISQW ILEFILRQPL DDNILNDLIN VLPLPNDNLN LKKALLVRKI ESEIANGSVT 60 EKILELMELI EELDHQEGII RDSVATKAAY CAVAVECTVK FLKSERAGSD KGKYFEAVRR 120 IWKRRIDLIE KVENVGVVSE ELWSWRDEIE AAFWEDRCAE NVIRRSKGVF AVDAVKAFVR 180 EAKERIGAPF LEVVAETYHT DEAVKSVFGE VNKGGASKKN GKEVCKGNAL PRKKHVAFKR 240 TRGASGIRIS DSIESEVEAS GGQDDSSAEV KIAKKALKLS SLELRAMVKD PLPDALRFAE 300 TLSSVVRDNM GHQPAENNSD MAPQPMVSSS RMAQAGGENR EAQQNCHHAA ASKPDLANRS 360 SAAHTFEWDS FLDDINEGSP SGVNRVTLPS PKRTKVSPLK KYEFKKITTR RKPVKWSALE 420 EDTLRTGVQK YGIGNWKVIL DAYRDIFTTR QSGDLKDKWR NMIS* |
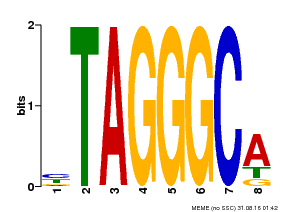
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00052 | PBM | Transfer from AT1G15720 | Download |
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_016577606.1 | 0.0 | PREDICTED: telomeric repeat-binding factor 2-like | ||||
| TrEMBL | A0A1U8H299 | 0.0 | A0A1U8H299_CAPAN; Uncharacterized protein | ||||
| TrEMBL | A0A2G2ZEE3 | 0.0 | A0A2G2ZEE3_CAPAN; telomeric repeat-binding factor 2-like | ||||
| STRING | PGSC0003DMT400078128 | 0.0 | (Solanum tuberosum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA3774 | 24 | 39 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G15720.1 | 1e-18 | TRF-like 5 | ||||



