 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | CA07g06670 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Solanoideae; Capsiceae; Capsicum
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 514aa MW: 56805.4 Da PI: 7.2323 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 54.2 | 2.7e-17 | 333 | 379 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
hn ErrRRdriN+++ L++llP++ +K +Ka++L +A+eY+ksLq
CA07g06670 333 VHNLSERRRRDRINEKMKALQDLLPHS-----TKTDKASMLDEAIEYLKSLQ 379
6*************************9.....9******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50888 | 18.501 | 329 | 378 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 4.06E-20 | 332 | 404 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 2.40E-18 | 332 | 383 | No hit | No description |
| Pfam | PF00010 | 1.1E-14 | 333 | 379 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 3.4E-20 | 333 | 387 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 2.6E-18 | 335 | 384 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009704 | Biological Process | de-etiolation | ||||
| GO:0010161 | Biological Process | red light signaling pathway | ||||
| GO:0010244 | Biological Process | response to low fluence blue light stimulus by blue low-fluence system | ||||
| GO:0010600 | Biological Process | regulation of auxin biosynthetic process | ||||
| GO:0010928 | Biological Process | regulation of auxin mediated signaling pathway | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 514 aa Download sequence Send to blast |
MNPCLPEWNI EAEQPVPHQK KPMGFDHELV ELLWRNGEVV LHSQTHKKQP GYDPNECRHF 60 DKHVQPTIRV AGDQTNLLQD DETVSWLNCP IDDSFDKVFC SPFLSDISTA PLREADKSIR 120 QSEDNKVFKF DPLEINHVLP QSHHSGFDPN PMPPPRFNNF GSAQQQHHIG GNQKGVNFPP 180 PVRSSNMQLG GKEARSNLML QDIKEGSVMT VGSSHCGSNQ VAVDADTSRF SSSANRGLSA 240 ALITDYTGKI SPQSDTMDRD TFEPANTSSS GGSGSSYARA CNRSTATNSQ NHKRKSRDGE 300 EPECQSKADG LESAGGNKAQ KSGTARRSRA AEVHNLSERR RRDRINEKMK ALQDLLPHST 360 KTDKASMLDE AIEYLKSLQM QLQMMWMGSG MASLMFPGVQ HYMSRMGMGI SPQTVPSIHN 420 AMHLARLPLV DPSIPLTQAA PNQAAMCQNS MLNQVNYQRH LQNPNFQDQY ASYMGFHPLQ 480 GASQPMNIFG LGSHTAQQSQ QLPHPTNSNA PTT* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 337 | 342 | ERRRRD |
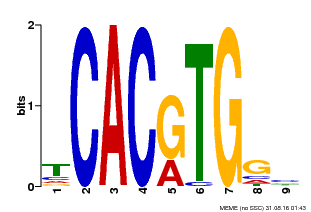
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00606 | ChIP-seq | Transfer from AT2G43010 | Download |
| |||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT013582 | 0.0 | BT013582.1 Lycopersicon esculentum clone 132330F, mRNA sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_016580439.1 | 0.0 | PREDICTED: transcription factor PIF4 | ||||
| TrEMBL | A0A1U8HIU3 | 0.0 | A0A1U8HIU3_CAPAN; Uncharacterized protein | ||||
| TrEMBL | A0A2G2Z1M5 | 0.0 | A0A2G2Z1M5_CAPAN; transcription factor PIF4 | ||||
| STRING | PGSC0003DMT400041156 | 0.0 | (Solanum tuberosum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA10465 | 22 | 26 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G59060.1 | 7e-44 | phytochrome interacting factor 3-like 6 | ||||



