 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | CCG000518.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Salicaceae; Saliceae; Populus
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 928aa MW: 104242 Da PI: 7.0165 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 160 | 4.5e-50 | 30 | 146 | 3 | 118 |
CG-1 3 ke.kkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptfqrrcy 99
+e ++rwl+++ei+a+L n++ +++ ++ + p sg++++++rk++r+frkDG++wkkkkdgktv+E+he+LKvg+ e +++yYah+++ ptf rrcy
CCG000518.1 30 EEsRTRWLRPNEIHAMLCNHKYFTINVKPVNLPMSGTIVFFDRKMLRNFRKDGHNWKKKKDGKTVKEAHEHLKVGNEERIHVYYAHGQDIPTFVRRCY 127
4459********************************************************************************************** PP
CG-1 100 wlLeeelekivlvhylevk 118
wlL+++le+ivlvhy+e++
CCG000518.1 128 WLLDKTLEHIVLVHYRETQ 146
****************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 74.993 | 25 | 151 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 1.8E-73 | 28 | 146 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 1.5E-44 | 32 | 144 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF01833 | 1.2E-6 | 378 | 464 | IPR002909 | IPT domain |
| SuperFamily | SSF81296 | 2.17E-14 | 379 | 465 | IPR014756 | Immunoglobulin E-set |
| Pfam | PF12796 | 2.8E-7 | 563 | 643 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 5.8E-16 | 563 | 676 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 1.06E-15 | 563 | 673 | No hit | No description |
| SuperFamily | SSF48403 | 2.8E-17 | 564 | 676 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 15.459 | 581 | 646 | IPR020683 | Ankyrin repeat-containing domain |
| SMART | SM00248 | 1.2E-6 | 614 | 643 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50088 | 11.915 | 614 | 646 | IPR002110 | Ankyrin repeat |
| SuperFamily | SSF52540 | 1.51E-6 | 725 | 828 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| SMART | SM00015 | 25 | 777 | 799 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 6.76 | 778 | 807 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 5.4E-4 | 800 | 822 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 10.109 | 801 | 825 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 3.5E-4 | 802 | 822 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 19 | 880 | 902 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 8.078 | 882 | 910 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 928 aa Download sequence Send to blast |
MESGYSDRLV GSEIHGFHTL QDLDVPSIME ESRTRWLRPN EIHAMLCNHK YFTINVKPVN 60 LPMSGTIVFF DRKMLRNFRK DGHNWKKKKD GKTVKEAHEH LKVGNEERIH VYYAHGQDIP 120 TFVRRCYWLL DKTLEHIVLV HYRETQELQG SPATPVNSHS SSVSDQSAPG LLSEESDSGA 180 ARGYYAGEKD LGPSDSLTVI NHAMRLHELN TLEWDDLLTN DPGNSILHGG DKIPSFDQQN 240 QIAVKGSVND GSTLSGYQLS AEKSALGNLT EAVVRNGNTQ FSGPDNVYRQ LTTDSQEYLD 300 AQRKNSVVLG AGDSLDILIN DGLQSQDSFG RWMNSIIDDS PVSVDDAVVE SSISSGYDSF 360 ASPGMDQHQS SIQEQMFIIT DFSPAWGFSN ETTKILVTGY FHEQYLHLAK SNLFCICGDA 420 FVPAEIVQAG VYSCMVSPHS PGLVNLCLSL DGSKPISQIL NFEYRAPLVH DSVVFSEVKS 480 KWEEFHLQMR LAYLLFSTSK TLNVLSSKVS PAKLKEAKKF AHRTSNISNS WAYLIKSIED 540 NRISVAQAKD GLFELSLKNT IKEWLLERVL EGCKTTEYDA QGLGVIHLCA IIGYTWAVYL 600 FSWSGLSLDF RDKHGWTALH WAAYYGREKM VAALLSAGAK PNLVTDPTKE NPGGCTAADL 660 ASAKGYDGLA AYLSEKALVA QFESMIIAGN ASGSLQTTAT DTVNSENLSE EELHLKDTLA 720 AYRTAADAAA RIQAAFREHS LKVYTKAVQS SSPEDEARNI IAAMKIQHAF RNYESKKKMA 780 AAAHIQHRFR TWKMRKNFLN MRRQAIKIQA AFRGFQVRKQ YRKIIWSVGL LEKAILRWRL 840 KRKGFRGLQV EPVETDVDPK HESDKEEDFY KISQKQAGER VERSVIRVQA MFRSKQAQEQ 900 YRRMKLTYNQ ATVEYEGLLD TDMVEGDL |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator (PubMed:14581622). Binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3' (By similarity). Regulates transcriptional activity in response to calcium signals (Probable). Binds calmodulin in a calcium-dependent manner (By similarity). Involved in response to cold. Contributes together with CAMTA3 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:28351986). {ECO:0000250|UniProtKB:Q8GSA7, ECO:0000269|PubMed:14581622, ECO:0000269|PubMed:28351986, ECO:0000305|PubMed:11925432}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00435 | DAP | Transfer from AT4G16150 | Download |
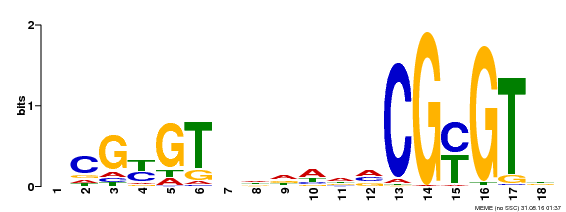
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By heat shock, UVB, wounding, abscisic acid, H(2)O(2) and salicylic acid. {ECO:0000269|PubMed:12218065}. | |||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AC208139 | 0.0 | AC208139.1 Populus trichocarpa clone JGIACSB13-D20, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011020359.1 | 0.0 | PREDICTED: calmodulin-binding transcription activator 5 isoform X3 | ||||
| Swissprot | O23463 | 0.0 | CMTA5_ARATH; Calmodulin-binding transcription activator 5 | ||||
| TrEMBL | A0A3N7FIW0 | 0.0 | A0A3N7FIW0_POPTR; Uncharacterized protein | ||||
| STRING | POPTR_0008s10730.1 | 0.0 | (Populus trichocarpa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF4288 | 34 | 59 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G16150.1 | 0.0 | calmodulin binding;transcription regulators | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



