 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | CCG002826.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Salicaceae; Saliceae; Populus
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 707aa MW: 77767.3 Da PI: 6.7811 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 61 | 1.9e-19 | 27 | 82 | 1 | 56 |
TT--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 1 rrkRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
+++ +++t+ q++ Le++F+++++p++++r +L+++lgL rq+k+WFqNrR+++k
CCG002826.1 27 KKRYHRHTALQIQKLESMFKECPHPDEKQRLQLSRELGLAPRQIKFWFQNRRTQMK 82
678899***********************************************998 PP
| |||||||
| 2 | START | 194.5 | 4.9e-61 | 221 | 442 | 2 | 206 |
HHHHHHHHHHHHHHHC-TT-EEEE....EXCCTTEEEEEEESSS.......SCEEEEEEEECCSCHHHHHHHHHCCCGGCT-TT-S....EEEEEEEE CS
START 2 laeeaaqelvkkalaeepgWvkss....esengdevlqkfeeskv......dsgealrasgvvdmvlallveellddkeqWdetla....kaetlevi 85
+a a++el+++ + +ep+W+kss + +n d++ ++f+++ + ++ea+r+sgvv+m+ + lv ++d++ +W e ++ a+t+evi
CCG002826.1 221 VAANAMEELLRLLQTNEPLWMKSSadgrDVLNLDTYQRIFPRAMShlknpnVRIEASRDSGVVIMNGVALVDMFMDSN-KWVESFPtmvsVAKTIEVI 317
67899***********************9***********998889999999**************************.99999999999******** PP
CTT......EEEEEEEEXXTTXX-SSX.EEEEEEEEEEE.TTS-EEEEEEEEE-TTS--.-TTSEE-EESSEEEEEEEECTCEEEEEEEE-EE--SSX CS
START 86 ssg......galqlmvaelqalsplvp.RdfvfvRyirqlgagdwvivdvSvdseqkppesssvvRaellpSgiliepksnghskvtwvehvdlkgrl 176
ssg g lqlm+ elq+lsplvp R+f ++Ry++q ++g w+iv+vS d +q+ ++ R+++lpSg+li++++ng+skvtwvehv+ ++++
CCG002826.1 318 SSGmlgshsGSLQLMYEELQVLSPLVPtREFCILRYCQQIEQGLWAIVSVSYDIPQFASQ----FRCHRLPSGCLIQDMPNGYSKVTWVEHVEIEDKT 411
*********************************************************987....*********************************9 PP
X.HHHHHHHHHHHHHHHHHHHHHHTXXXXXX CS
START 177 p.hwllrslvksglaegaktwvatlqrqcek 206
h+l+r l++sg+a+ga +w+atlqr ce+
CCG002826.1 412 TtHQLYRDLIHSGMAFGAERWLATLQRMCER 442
99***************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 2.05E-20 | 10 | 84 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 1.3E-22 | 11 | 78 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.815 | 24 | 84 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 1.8E-18 | 25 | 88 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 2.84E-18 | 26 | 85 | No hit | No description |
| Pfam | PF00046 | 5.1E-17 | 27 | 82 | IPR001356 | Homeobox domain |
| PROSITE pattern | PS00027 | 0 | 59 | 82 | IPR017970 | Homeobox, conserved site |
| PROSITE profile | PS50848 | 51.974 | 211 | 445 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 6.32E-39 | 212 | 443 | No hit | No description |
| CDD | cd08875 | 6.06E-120 | 215 | 441 | No hit | No description |
| SMART | SM00234 | 1.6E-54 | 220 | 442 | IPR002913 | START domain |
| Pfam | PF01852 | 1.7E-53 | 221 | 442 | IPR002913 | START domain |
| Gene3D | G3DSA:3.30.530.20 | 9.0E-9 | 288 | 427 | IPR023393 | START-like domain |
| SuperFamily | SSF55961 | 8.52E-23 | 463 | 699 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009828 | Biological Process | plant-type cell wall loosening | ||||
| GO:0010091 | Biological Process | trichome branching | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0000976 | Molecular Function | transcription regulatory region sequence-specific DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008289 | Molecular Function | lipid binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 707 aa Download sequence Send to blast |
MEYGSVGGGN SGGGGGDHDP SDPQRRKKRY HRHTALQIQK LESMFKECPH PDEKQRLQLS 60 RELGLAPRQI KFWFQNRRTQ MKAQHERADN SALRAENDKI RCENIAIREA LKNVICPSCG 120 GPPVTEDSYF DEHKLRMENV QLKEELDRVS SIAAKYVGRP ISQLPPVQPF HFSSLDLSMG 180 NFGGQGIGGP SLDLDLIPTS SNLAFQPPVI SDMDKSLMTD VAANAMEELL RLLQTNEPLW 240 MKSSADGRDV LNLDTYQRIF PRAMSHLKNP NVRIEASRDS GVVIMNGVAL VDMFMDSNKW 300 VESFPTMVSV AKTIEVISSG MLGSHSGSLQ LMYEELQVLS PLVPTREFCI LRYCQQIEQG 360 LWAIVSVSYD IPQFASQFRC HRLPSGCLIQ DMPNGYSKVT WVEHVEIEDK TTTHQLYRDL 420 IHSGMAFGAE RWLATLQRMC ERVACQMVSS NSTRDLGGVI PSPEGKRSMM KLAQRMVSSF 480 CSSISTSNSH RWSTLSGLHD VGVRVTLHKS TDPGQPNGVV LSAATTFSLP VSPQNVFSFF 540 KDERTRPQWD VLSSGNAVQE VAHITNGSHP GNCISVLRAY NTSQNNMLIL QESCVDSSGS 600 LVVYCPVDLP AINIAMSGED PSYIPLLPSG FTISPDGRPD QGDGASTSSN TQGSTARLSG 660 SLITVAFQIL VSSLPSAKLN LESVNTVNNL IGTTVQQIKA AMNCPSS |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor which acts as positive regulator of drought stress tolerance. Can transactivate CIPK3, NCED3 and ERECTA (PubMed:18451323). Transactivates several cell-wall-loosening protein genes by directly binding to HD motifs in their promoters. These target genes play important roles in coordinating cell-wall extensibility with root development and growth (PubMed:24821957). Transactivates CYP74A/AOS, AOC3, OPR3 and 4CLL5/OPCL1 genes by directly binding to HD motifs in their promoters. These target genes are involved in jasmonate (JA) biosynthesis, and JA signaling affects root architecture by activating auxin signaling, which promotes lateral root formation (PubMed:25752924). Acts as negative regulator of trichome branching (PubMed:16778018, PubMed:24824485). Required for the establishment of giant cell identity on the abaxial side of sepals (PubMed:23095885). May regulate cell differentiation and proliferation during root and shoot meristem development (PubMed:25564655). {ECO:0000269|PubMed:16778018, ECO:0000269|PubMed:18451323, ECO:0000269|PubMed:23095885, ECO:0000269|PubMed:24821957, ECO:0000269|PubMed:24824485, ECO:0000269|PubMed:25564655, ECO:0000269|PubMed:25752924}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00232 | DAP | Transfer from AT1G73360 | Download |
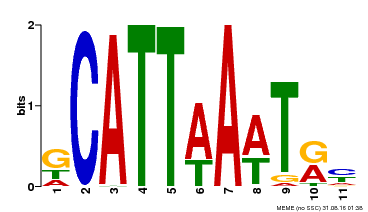
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011035552.1 | 0.0 | PREDICTED: homeobox-leucine zipper protein HDG11 | ||||
| Swissprot | Q9FX31 | 0.0 | HDG11_ARATH; Homeobox-leucine zipper protein HDG11 | ||||
| TrEMBL | B9N3B2 | 0.0 | B9N3B2_POPTR; Uncharacterized protein | ||||
| STRING | POPTR_0015s05050.1 | 0.0 | (Populus trichocarpa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1026 | 33 | 98 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G73360.1 | 0.0 | homeodomain GLABROUS 11 | ||||



